Gene
KWMTBOMO07303
Pre Gene Modal
BGIBMGA010658
Annotation
PREDICTED:_cleft_lip_and_palate_transmembrane_protein_1_homolog_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.931
Sequence
CDS
ATGTCTGAAGCTGAAAAAATGTTATCTACTCAGTCTGGTCCTCCTGAAAATGAATCATCAGAGAACTCTGAAAACATAAGCCACGATGGTTCAAGTGTGGTTGCTGCAGATGATAATCTCGATATTAATGCTCGCGTTGAAGAGGAACGTCGTAGAATGCAACCAACTAAATTAGAATCATTCTTTGCCATTACAAAATCATTGATAATCAGAGCTTTAGTAGTTTATTTTATTACTTCTATGTTTAGACAGCCATCTGCTCCAAAAACAGATATCAATAATCCATCTAGTGCCCCAAGAGCACCAGCTGTGAATATGTTTGCAAATGGAACTATACTAGACATGTACTGTTATCTGTCTGAACAAGAATTTAATACAAATTTTGACAATTCAAATTTAATTTGGCAGCACAGTGGTTTGGTGTATGGAGATTGGTATTCAGGTCCGAATGGTGATGGATCATACAGTCACTCAGCAAGCATAACACCATCATTTGCGTTAAAAAATAATGGATCCATTTATCTACATGTTTATATTGTACCTTCTGGGAAATCACCGGATCCTAACAATAGACAAAACTTTGCTGGCCCTTACATCTCTGTTGAGAAAAAGATGTTAAATAAATATAAGAAGTTGAAGTATCAAAAGACTCTAAATTTACTAACAGGTCAAACAGAGAAATCTGAAGAAGAAATTAAGAAAGCTGAAACTGTAAAAGAAGAAATTGTTTCACATTGGCATCCTAACTTAACTATTAATTTGGTAAATGACCAAACAAATTGGGTACAAGGGAGTGTACCACCACCTCTAGATGAATTCATTTATTTTCTACCTGATGGTAAACAATATAAACCTGCTGTATTTTTAAATGATTACTGGAATATGATGCGTGATTATGTACCTATTAATAAAACAACAGAAGTTCTTAACTTACAATTGACATATCAACCACTTAGTCTGTTCAAGTGGCAACTTTATACTGCACAGGCAATGAGAGACAAGCTTAGCATGTTTTCTGCTTTAGGGGCTGAAGAACAGGATGAAGACCAGGACACTGTAAAGGAATTGTTACTTGACACTTCTCCATATTTGCTGGCATTGACAATCTCAGTTTCAATTTTACACTCTATATGTGAACTTTTAGCATTCAAAAATGATATACAGTTTTGGAACAACAGGCAGTCTTTAGAAGGATTATCTGTCCGTTCAGTATTTTTTAATGTTTTCCAGTCTACAATTGTACTCCTCTATGTTCTGGATAATGAAACCAATGTAATGGTTAGAATATCGTGTTTCATTGGATTATTAATTGAAATTTGGAAAATAAATAAAGTTATGGATGTTAAGATAAATAGAGAAAGCCGTATACTGGGCATTCCAAAACTGTCATTTACTGACAAGGGATCATATGTGGAATCTAGCACTAAACAATATGACATGTTGGCTTTTCGTTATTTGAGTTGGGCATGTTTTCCACTCTTGATTGGCTATGGTGTTTATTCATTACTGTATCAGGAACATAAAGGGTGGTATTCCTTCATATTGAACATGATGTATGGCTATCTTTTGACATTTGGATTCATTATGATGACCCCACAATTATTCATCAATTATAAGTTAAAATCGGTGGCCCATCTTCCTTGGCGAATGATGACATACAAATTTCTGAACACATTCATTGATGATATTTTTGCATTTGTAATCAAAATGCCAACTATGTATAGATTGGGATGTTTTAGAGATGATATAGTATTCTTTATCTTTTTGTACCAACGCTGGATTTACAAGGTTGACCACAAACGTGTTAATGAATTTGGCTTTTCTGGTGAAATGGAAGAGCAACAGAAGCAAGCTAAAAATGGAGCTCTAGCTATTACTGAATCTGAAGATATTGCCAACAAGAAGAATGATTAA
Protein
MSEAEKMLSTQSGPPENESSENSENISHDGSSVVAADDNLDINARVEEERRRMQPTKLESFFAITKSLIIRALVVYFITSMFRQPSAPKTDINNPSSAPRAPAVNMFANGTILDMYCYLSEQEFNTNFDNSNLIWQHSGLVYGDWYSGPNGDGSYSHSASITPSFALKNNGSIYLHVYIVPSGKSPDPNNRQNFAGPYISVEKKMLNKYKKLKYQKTLNLLTGQTEKSEEEIKKAETVKEEIVSHWHPNLTINLVNDQTNWVQGSVPPPLDEFIYFLPDGKQYKPAVFLNDYWNMMRDYVPINKTTEVLNLQLTYQPLSLFKWQLYTAQAMRDKLSMFSALGAEEQDEDQDTVKELLLDTSPYLLALTISVSILHSICELLAFKNDIQFWNNRQSLEGLSVRSVFFNVFQSTIVLLYVLDNETNVMVRISCFIGLLIEIWKINKVMDVKINRESRILGIPKLSFTDKGSYVESSTKQYDMLAFRYLSWACFPLLIGYGVYSLLYQEHKGWYSFILNMMYGYLLTFGFIMMTPQLFINYKLKSVAHLPWRMMTYKFLNTFIDDIFAFVIKMPTMYRLGCFRDDIVFFIFLYQRWIYKVDHKRVNEFGFSGEMEEQQKQAKNGALAITESEDIANKKND
Summary
Uniprot
A0A2A4J749
A0A2H1VG74
A0A3S2LPA9
A0A0N1IQE6
A0A194PJ06
A0A0L7LSZ4
+ More
H9JMA5 A0A1E1WNT2 E2AUW1 U5EUK0 A0A182QM45 B0W5N9 A0A1B0D4J5 A0A2J7PS95 A0A023EVJ6 A0A182NE37 A0A182GBS7 E2B8D0 A0A2P8YIS7 A0A182GR85 A0A1B6G4Q6 A0A195FJY0 A0A336LTZ5 A0A1Q3F8A7 A0A088ANA5 A0A1Q3F8K8 V9ILG1 A0A0M8ZQZ8 A0A2A3EBS5 K7IVY1 A0A026W5S9 A0A1L8D916 A0A1S4FDT7 A0A195BFE7 A0A2M4BG69 A0A158NRB2 Q0IF41 A0A084VP32 Q7Q3K6 A0A1J1HNG3 A0A151JBI8 A0A182MPU0 A0A182K1E8 A0A2M3Z1P9 A0A182WT85 A0A182YGU4 A0A1Y9G8B5 A0A182W7V6 A0A182HV78 A0A182V2N2 A0A1S4GWP2 A0A182U551 A0A182L230 A0A232EVH6 W5J870 A0A182J1B6 A0A151IFL4 A0A2M4AE74 A0A0L7RGT4 A0A182RBL9 A0A0A1XN51 A0A182PJ72 W8C2I6 A0A034WSI4 A0A034WNK1 A0A1J1IMS4 A0A3B0J9Y8 A0A0K8VWI2 A0A0C9QTJ5 B3MLU7 B4N0N2 A0A0K8TQ32 A0A1I8N9P0 B4NYI3 B3N3P9 A0A1W4V2Y1 B4KFP8 A0A1I8QDB9 A0A0L0CBR7 A0A1B6E9K7 T1PDI7 B4MDC7 Q29NP2 B4GQR0 B4JD39 A0A240SWL2 A0A1B0B5R7 A0A1A9UMV9 A0A0M5IVY0 A0A1A9ZJ31 B4I370 Q9VQX6 A0A1W7RAS7 B4NRW3 A0A0N7Z9A4 T1HZ64 A0A1A9W0F4 R4G484 A0A087UJA7
H9JMA5 A0A1E1WNT2 E2AUW1 U5EUK0 A0A182QM45 B0W5N9 A0A1B0D4J5 A0A2J7PS95 A0A023EVJ6 A0A182NE37 A0A182GBS7 E2B8D0 A0A2P8YIS7 A0A182GR85 A0A1B6G4Q6 A0A195FJY0 A0A336LTZ5 A0A1Q3F8A7 A0A088ANA5 A0A1Q3F8K8 V9ILG1 A0A0M8ZQZ8 A0A2A3EBS5 K7IVY1 A0A026W5S9 A0A1L8D916 A0A1S4FDT7 A0A195BFE7 A0A2M4BG69 A0A158NRB2 Q0IF41 A0A084VP32 Q7Q3K6 A0A1J1HNG3 A0A151JBI8 A0A182MPU0 A0A182K1E8 A0A2M3Z1P9 A0A182WT85 A0A182YGU4 A0A1Y9G8B5 A0A182W7V6 A0A182HV78 A0A182V2N2 A0A1S4GWP2 A0A182U551 A0A182L230 A0A232EVH6 W5J870 A0A182J1B6 A0A151IFL4 A0A2M4AE74 A0A0L7RGT4 A0A182RBL9 A0A0A1XN51 A0A182PJ72 W8C2I6 A0A034WSI4 A0A034WNK1 A0A1J1IMS4 A0A3B0J9Y8 A0A0K8VWI2 A0A0C9QTJ5 B3MLU7 B4N0N2 A0A0K8TQ32 A0A1I8N9P0 B4NYI3 B3N3P9 A0A1W4V2Y1 B4KFP8 A0A1I8QDB9 A0A0L0CBR7 A0A1B6E9K7 T1PDI7 B4MDC7 Q29NP2 B4GQR0 B4JD39 A0A240SWL2 A0A1B0B5R7 A0A1A9UMV9 A0A0M5IVY0 A0A1A9ZJ31 B4I370 Q9VQX6 A0A1W7RAS7 B4NRW3 A0A0N7Z9A4 T1HZ64 A0A1A9W0F4 R4G484 A0A087UJA7
Pubmed
26354079
26227816
19121390
20798317
24945155
26483478
+ More
29403074 20075255 24508170 30249741 21347285 17510324 24438588 12364791 25244985 20966253 28648823 20920257 23761445 25830018 24495485 25348373 17994087 26369729 25315136 17550304 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 27129103
29403074 20075255 24508170 30249741 21347285 17510324 24438588 12364791 25244985 20966253 28648823 20920257 23761445 25830018 24495485 25348373 17994087 26369729 25315136 17550304 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 27129103
EMBL
NWSH01002640
PCG67781.1
ODYU01002368
SOQ39781.1
RSAL01000504
RVE41518.1
+ More
KQ459752 KPJ20202.1 KQ459603 KPI92704.1 JTDY01000153 KOB78567.1 BABH01033372 BABH01033373 GDQN01002394 JAT88660.1 GL442898 EFN62769.1 GANO01002284 JAB57587.1 AXCN02000872 DS231844 EDS35692.1 AJVK01003266 NEVH01021938 PNF19201.1 GAPW01000420 JAC13178.1 JXUM01052982 KQ561757 KXJ77620.1 GL446310 EFN88050.1 PYGN01000568 PSN44104.1 JXUM01016288 JXUM01016289 JXUM01016290 KQ560454 KXJ82338.1 GECZ01012350 JAS57419.1 KQ981522 KYN40567.1 UFQT01000108 SSX20109.1 GFDL01011240 JAV23805.1 GFDL01011192 JAV23853.1 JR053561 AEY61963.1 KQ435944 KOX68196.1 KZ288292 PBC29187.1 KK107390 QOIP01000007 EZA51425.1 RLU20428.1 GFDF01011133 JAV02951.1 KQ976504 KYM82912.1 GGFJ01002915 MBW52056.1 ADTU01023845 CH477394 EAT41898.1 ATLV01014972 KE524999 KFB39726.1 AAAB01008964 EAA12218.3 CVRI01000013 CRK89589.1 KQ979122 KYN22521.1 AXCM01007613 GGFM01001617 MBW22368.1 APCN01002500 NNAY01001995 OXU22354.1 ADMH02001962 ETN60161.1 KQ977771 KYM99988.1 GGFK01005691 MBW39012.1 KQ414596 KOC70043.1 GBXI01001533 JAD12759.1 GAMC01006529 JAC00027.1 GAKP01001807 JAC57145.1 GAKP01001806 JAC57146.1 CVRI01000055 CRL01464.1 OUUW01000004 SPP79094.1 GDHF01009081 JAI43233.1 GBYB01003992 JAG73759.1 CH902620 EDV30818.1 CH963920 EDW77645.1 GDAI01001106 JAI16497.1 CM000157 EDW87616.1 CH954177 EDV57708.1 CH933807 EDW11013.1 JRES01000736 KNC28879.1 GEDC01002754 JAS34544.1 KA646826 AFP61455.1 CH940661 EDW71188.1 CH379059 EAL34176.1 CH479187 EDW39932.1 CH916368 EDW04283.1 JXJN01008839 CP012523 ALC38081.1 CH480820 EDW54215.1 AE014134 AY052040 AAF51035.2 AAK93464.1 GFAH01000152 JAV48237.1 CH981594 CM002910 EDX15341.1 KMY88034.1 GDKW01001099 JAI55496.1 ACPB03008638 GAHY01000996 JAA76514.1 KK120066 KFM77446.1
KQ459752 KPJ20202.1 KQ459603 KPI92704.1 JTDY01000153 KOB78567.1 BABH01033372 BABH01033373 GDQN01002394 JAT88660.1 GL442898 EFN62769.1 GANO01002284 JAB57587.1 AXCN02000872 DS231844 EDS35692.1 AJVK01003266 NEVH01021938 PNF19201.1 GAPW01000420 JAC13178.1 JXUM01052982 KQ561757 KXJ77620.1 GL446310 EFN88050.1 PYGN01000568 PSN44104.1 JXUM01016288 JXUM01016289 JXUM01016290 KQ560454 KXJ82338.1 GECZ01012350 JAS57419.1 KQ981522 KYN40567.1 UFQT01000108 SSX20109.1 GFDL01011240 JAV23805.1 GFDL01011192 JAV23853.1 JR053561 AEY61963.1 KQ435944 KOX68196.1 KZ288292 PBC29187.1 KK107390 QOIP01000007 EZA51425.1 RLU20428.1 GFDF01011133 JAV02951.1 KQ976504 KYM82912.1 GGFJ01002915 MBW52056.1 ADTU01023845 CH477394 EAT41898.1 ATLV01014972 KE524999 KFB39726.1 AAAB01008964 EAA12218.3 CVRI01000013 CRK89589.1 KQ979122 KYN22521.1 AXCM01007613 GGFM01001617 MBW22368.1 APCN01002500 NNAY01001995 OXU22354.1 ADMH02001962 ETN60161.1 KQ977771 KYM99988.1 GGFK01005691 MBW39012.1 KQ414596 KOC70043.1 GBXI01001533 JAD12759.1 GAMC01006529 JAC00027.1 GAKP01001807 JAC57145.1 GAKP01001806 JAC57146.1 CVRI01000055 CRL01464.1 OUUW01000004 SPP79094.1 GDHF01009081 JAI43233.1 GBYB01003992 JAG73759.1 CH902620 EDV30818.1 CH963920 EDW77645.1 GDAI01001106 JAI16497.1 CM000157 EDW87616.1 CH954177 EDV57708.1 CH933807 EDW11013.1 JRES01000736 KNC28879.1 GEDC01002754 JAS34544.1 KA646826 AFP61455.1 CH940661 EDW71188.1 CH379059 EAL34176.1 CH479187 EDW39932.1 CH916368 EDW04283.1 JXJN01008839 CP012523 ALC38081.1 CH480820 EDW54215.1 AE014134 AY052040 AAF51035.2 AAK93464.1 GFAH01000152 JAV48237.1 CH981594 CM002910 EDX15341.1 KMY88034.1 GDKW01001099 JAI55496.1 ACPB03008638 GAHY01000996 JAA76514.1 KK120066 KFM77446.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000037510
UP000005204
+ More
UP000000311 UP000075886 UP000002320 UP000092462 UP000235965 UP000075884 UP000069940 UP000249989 UP000008237 UP000245037 UP000078541 UP000005203 UP000053105 UP000242457 UP000002358 UP000053097 UP000279307 UP000078540 UP000005205 UP000008820 UP000030765 UP000007062 UP000183832 UP000078492 UP000075883 UP000075881 UP000076407 UP000076408 UP000069272 UP000075920 UP000075840 UP000075903 UP000075902 UP000075882 UP000215335 UP000000673 UP000075880 UP000078542 UP000053825 UP000075900 UP000075885 UP000268350 UP000007801 UP000007798 UP000095301 UP000002282 UP000008711 UP000192221 UP000009192 UP000095300 UP000037069 UP000008792 UP000001819 UP000008744 UP000001070 UP000092443 UP000092460 UP000078200 UP000092553 UP000092445 UP000001292 UP000000803 UP000000304 UP000015103 UP000091820 UP000054359
UP000000311 UP000075886 UP000002320 UP000092462 UP000235965 UP000075884 UP000069940 UP000249989 UP000008237 UP000245037 UP000078541 UP000005203 UP000053105 UP000242457 UP000002358 UP000053097 UP000279307 UP000078540 UP000005205 UP000008820 UP000030765 UP000007062 UP000183832 UP000078492 UP000075883 UP000075881 UP000076407 UP000076408 UP000069272 UP000075920 UP000075840 UP000075903 UP000075902 UP000075882 UP000215335 UP000000673 UP000075880 UP000078542 UP000053825 UP000075900 UP000075885 UP000268350 UP000007801 UP000007798 UP000095301 UP000002282 UP000008711 UP000192221 UP000009192 UP000095300 UP000037069 UP000008792 UP000001819 UP000008744 UP000001070 UP000092443 UP000092460 UP000078200 UP000092553 UP000092445 UP000001292 UP000000803 UP000000304 UP000015103 UP000091820 UP000054359
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J749
A0A2H1VG74
A0A3S2LPA9
A0A0N1IQE6
A0A194PJ06
A0A0L7LSZ4
+ More
H9JMA5 A0A1E1WNT2 E2AUW1 U5EUK0 A0A182QM45 B0W5N9 A0A1B0D4J5 A0A2J7PS95 A0A023EVJ6 A0A182NE37 A0A182GBS7 E2B8D0 A0A2P8YIS7 A0A182GR85 A0A1B6G4Q6 A0A195FJY0 A0A336LTZ5 A0A1Q3F8A7 A0A088ANA5 A0A1Q3F8K8 V9ILG1 A0A0M8ZQZ8 A0A2A3EBS5 K7IVY1 A0A026W5S9 A0A1L8D916 A0A1S4FDT7 A0A195BFE7 A0A2M4BG69 A0A158NRB2 Q0IF41 A0A084VP32 Q7Q3K6 A0A1J1HNG3 A0A151JBI8 A0A182MPU0 A0A182K1E8 A0A2M3Z1P9 A0A182WT85 A0A182YGU4 A0A1Y9G8B5 A0A182W7V6 A0A182HV78 A0A182V2N2 A0A1S4GWP2 A0A182U551 A0A182L230 A0A232EVH6 W5J870 A0A182J1B6 A0A151IFL4 A0A2M4AE74 A0A0L7RGT4 A0A182RBL9 A0A0A1XN51 A0A182PJ72 W8C2I6 A0A034WSI4 A0A034WNK1 A0A1J1IMS4 A0A3B0J9Y8 A0A0K8VWI2 A0A0C9QTJ5 B3MLU7 B4N0N2 A0A0K8TQ32 A0A1I8N9P0 B4NYI3 B3N3P9 A0A1W4V2Y1 B4KFP8 A0A1I8QDB9 A0A0L0CBR7 A0A1B6E9K7 T1PDI7 B4MDC7 Q29NP2 B4GQR0 B4JD39 A0A240SWL2 A0A1B0B5R7 A0A1A9UMV9 A0A0M5IVY0 A0A1A9ZJ31 B4I370 Q9VQX6 A0A1W7RAS7 B4NRW3 A0A0N7Z9A4 T1HZ64 A0A1A9W0F4 R4G484 A0A087UJA7
H9JMA5 A0A1E1WNT2 E2AUW1 U5EUK0 A0A182QM45 B0W5N9 A0A1B0D4J5 A0A2J7PS95 A0A023EVJ6 A0A182NE37 A0A182GBS7 E2B8D0 A0A2P8YIS7 A0A182GR85 A0A1B6G4Q6 A0A195FJY0 A0A336LTZ5 A0A1Q3F8A7 A0A088ANA5 A0A1Q3F8K8 V9ILG1 A0A0M8ZQZ8 A0A2A3EBS5 K7IVY1 A0A026W5S9 A0A1L8D916 A0A1S4FDT7 A0A195BFE7 A0A2M4BG69 A0A158NRB2 Q0IF41 A0A084VP32 Q7Q3K6 A0A1J1HNG3 A0A151JBI8 A0A182MPU0 A0A182K1E8 A0A2M3Z1P9 A0A182WT85 A0A182YGU4 A0A1Y9G8B5 A0A182W7V6 A0A182HV78 A0A182V2N2 A0A1S4GWP2 A0A182U551 A0A182L230 A0A232EVH6 W5J870 A0A182J1B6 A0A151IFL4 A0A2M4AE74 A0A0L7RGT4 A0A182RBL9 A0A0A1XN51 A0A182PJ72 W8C2I6 A0A034WSI4 A0A034WNK1 A0A1J1IMS4 A0A3B0J9Y8 A0A0K8VWI2 A0A0C9QTJ5 B3MLU7 B4N0N2 A0A0K8TQ32 A0A1I8N9P0 B4NYI3 B3N3P9 A0A1W4V2Y1 B4KFP8 A0A1I8QDB9 A0A0L0CBR7 A0A1B6E9K7 T1PDI7 B4MDC7 Q29NP2 B4GQR0 B4JD39 A0A240SWL2 A0A1B0B5R7 A0A1A9UMV9 A0A0M5IVY0 A0A1A9ZJ31 B4I370 Q9VQX6 A0A1W7RAS7 B4NRW3 A0A0N7Z9A4 T1HZ64 A0A1A9W0F4 R4G484 A0A087UJA7
Ontologies
GO
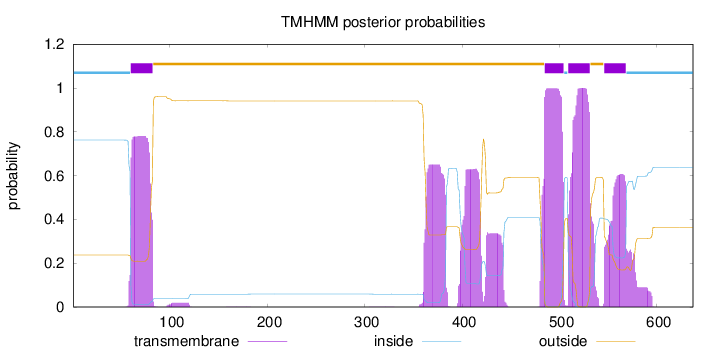
Topology
Length:
637
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
109.2664
Exp number, first 60 AAs:
0.95345
Total prob of N-in:
0.76239
inside
1 - 59
TMhelix
60 - 82
outside
83 - 484
TMhelix
485 - 504
inside
505 - 508
TMhelix
509 - 531
outside
532 - 545
TMhelix
546 - 568
inside
569 - 637
Population Genetic Test Statistics
Pi
6.81691
Theta
6.530325
Tajima's D
0.7196
CLR
0.637386
CSRT
0.59722013899305
Interpretation
Uncertain
