Gene
KWMTBOMO07302 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010657
Annotation
CRM1_[Spodoptera_frugiperda]
Location in the cell
Cytoplasmic Reliability : 1.273 PlasmaMembrane Reliability : 1.628
Sequence
CDS
ATGGCAACCTTAGAACAACAGGCATCTAAACTTTTGGATTTCAATCAAAAATTGGACATCAATTTATTGGACAACATTGTTGGCTGTTTATATTCTGCTGTTGGGGATCAACAACGTGTTGCTCAGGATATACTGACTGCTTTGAAGGAACATCCTGATGCTTGGACTCGTGTTGATACAATACTTGAATATTCTCAAAATCAAGAAACTAAATATTATGCATTACAAATATTAGAACAAGTAATACTAACGAGATGGAAAATATTACCAAGAAATCAGTGTGAGGGTATAAAAAAATATATAGTTGGCCTTATCATTAAAAATTCATCAGATCCTGTTACGATGGAAAGCAACAAAGTTTATTTGAAAAAACTAAACATGATCCTTATTCAAGTTCTGAAAAGAGAATGGCCCCATAATTGGGAAACTTTTATCAGTGACATTGTGGGTGCCTCAAAGACGAATGAAAGTCTCTGTCAAAACAATATGGAAATATTCAAGTTGTTGAGTGAAGAAGTATTTATGTTTAGTACAGGCCAGCTCACACAAACAAAAGCTAAGCATCTAAAAGATACAATGTGTTCAGAATTCAGTCAAATCTTTCAGCTATGCCAATTTGTCTTGGAAAATTCACAAAATGCACCTTTAGTTGGAGTGACCTTAGACACTCTTTTGAGATTTTTAAATTGGATTCCACTTGGTTATATTTTCGAAATGAAATTGATAAGCACATTGATATTTAAGTTTCTGAACGTGCCGATGTTTAGGAATGTAACATTGAGCTGTCTTACTGAAATTGCCGGTGTCACAGTCAGCAATTATGATCAGCAGTTTGTTTATTTGTTTGTACAAACTATGGAACAACTTGAAGTTATGCTTCCTCTAACTACAAACATACGTGAAGCATATGTAGCTGGTCGAGATCAGGAACAAGTTTTTATTCAAAATCTTGCACTTTTCTTATGCACATATCTAAAAGAGCATGGTCCGCTCATAGAAAAGAGTGGATTAACAAACACTTTAATGAATGGTTTGAGGTATCTTGTCTTAATATCAGAGGTAGAAGATGTAGAAATTTTCAAAATTTGTCTTGAATATTGGAATGCTCTTGCTGCTGATTTGTACAAAATTGCGCCATGTTCGTCAAACTTGTACCATTTGGGTAAAAGTGGAGGCCGAAAAGCTTTGTATGCTGATGTCTTGAGCAGTGTTCGGTATATTATGATCTCGAGAATGGCAAAACCAGAAGAGGTGTTAGTTGTTGAAAATGAAAATGGTGAAGTTGTGAGAGAATTTATGAAGGACACAGACTCCATTAATTTGTATAAAAATATGAGAGAAACATTAGTTTACTTGACACATTTAGACTATTTAGATACTGAACGTATTATGACAGAGAAATTACAAAATCAAGTAAATGGTACTGAGTGGTCATGGAAGAACTTGAACACATTATGTTGGGCAATTGGATCTATTTCTGGAGCTCTATCTGAGGATGATGAAAAGAGATTTTTGGTTATTGTGATTAAAGAGCTTCTAGGTCTCTGTGAACAGAAAAAGGGAAAAGATAACAAAGCTATAATTGCTAGTAACATAATGTATGTCGTAGGACAATACCCACGCTTTCTTCGTGCACATTGGAAATTTTTAAAAACCGTTGTCAATAAACTTTTTGAATTCATGCATGAAACACATGATGGTGTCCAAGACATGGCGTGTGATACATTCATAAAAATTGCTATAAAATGCCGTCGACATTTTGTCACTACTCAAGTTGGAGAAGCCTGTCCTTTTATTGAAGAAATATTGGGCACCATTAGTACTATAATTTGTGATTTACAAACATTGCAAGTTCACACTTTTTATGAAGCTGTTGGATACATGATCAGTGCCCAAGTAGATCAAGTAGCCCAAGAACTTTTAATTGAAAAATACATGCTTCTGCCTAATCAAGTTTGGGATGATATTATATCTCAAGCTTCTCGTAATGTTGAAATCTTGAAAGACCCTGAAGCTGTTAAACAACTTGTAAGTATACTGAAAACAAATGTGCGTGCATGTCGTGCACTAGCACACCCATATGTTGTGCAATTAGGAAGGATATACCTTGACATGCTTAATGTTTACAAAGTAATGTCTGAAAATATAAGTCAAGCCATAGCTTTAAATGGCGTAGCAGTTACTAAACAACCTCTCATTAAGAATATGAGAATAATAAAAAAGGAAACTTTGAACCTTATTTCAAGTTGGGTTACACGCTCGATTGATAACAGTATGGTTTTGGAAAACTTTATTCCACCTCTTCTCGATGCTGTGCTTTTGGATTACCAAAGGACTGCTGTTCCTGAAGCTCGTGAGCCTGAAGTACTGTCTTGCATGGCAGCAATCGTGCACAGACTTGAAGGACATATAACTTCTGAAGTACCAAAAATATTTGATGCAGTCTTTGAATGTACTCTGGAAATGATTAACAAAGACTTTGAAGAGTATCCAGAACATAGAACAGAATTTTTCTTATTACTGCAGGCGGTCAATACAAACTGTTTCAAAGCATTTTTGAGCATACCACCAGCACAATTCAAGTTGGTTTTAGATTCCATAATCTGGGCATTTAAACATACAATGAGGAATGTGGCAGATACCGGTTTACAAATATTGTACCGACTGTTGTTAAATGTTGAGGAACATCCACAGGCAGCACAGAGTTTTTACAGAACCTACCTTTGTGACATTTTGGAGCATGTCTTCAGTGTAGTCACTGATACATCACATGGTGCAGGACTCACTATGCACGCTACAATCCTGGCTCACATTTTCTCATTGGTAGAGGCTGGCCGAGTTACTGTACCACTTGGACCCACACCAGACAATGTTCTTTATATTCAGGAATATATCGCAAATCTCTTGAAAACAGCATTTTCACACTTAACTGATAACCAAATTAAAATTACTGTACAAGGCCTGTTCAACCTGAATCAAGATATACCTGCTTTCAAGGATCATCTCAGAGACTTCTTAGTTCAAATTAGAGAGTACACTGGTGAAGATGATAGCGATTTGTACTTGGAAGAGCGTTTGTTTGTATTACGTCAGGCACAAGAAGATAAGAGAACAGTGCAGTTGTCAGTGCCCGGCATTCTGAACCCTCATGAATTGCCTGAAGAGATGCAAGATTAA
Protein
MATLEQQASKLLDFNQKLDINLLDNIVGCLYSAVGDQQRVAQDILTALKEHPDAWTRVDTILEYSQNQETKYYALQILEQVILTRWKILPRNQCEGIKKYIVGLIIKNSSDPVTMESNKVYLKKLNMILIQVLKREWPHNWETFISDIVGASKTNESLCQNNMEIFKLLSEEVFMFSTGQLTQTKAKHLKDTMCSEFSQIFQLCQFVLENSQNAPLVGVTLDTLLRFLNWIPLGYIFEMKLISTLIFKFLNVPMFRNVTLSCLTEIAGVTVSNYDQQFVYLFVQTMEQLEVMLPLTTNIREAYVAGRDQEQVFIQNLALFLCTYLKEHGPLIEKSGLTNTLMNGLRYLVLISEVEDVEIFKICLEYWNALAADLYKIAPCSSNLYHLGKSGGRKALYADVLSSVRYIMISRMAKPEEVLVVENENGEVVREFMKDTDSINLYKNMRETLVYLTHLDYLDTERIMTEKLQNQVNGTEWSWKNLNTLCWAIGSISGALSEDDEKRFLVIVIKELLGLCEQKKGKDNKAIIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHETHDGVQDMACDTFIKIAIKCRRHFVTTQVGEACPFIEEILGTISTIICDLQTLQVHTFYEAVGYMISAQVDQVAQELLIEKYMLLPNQVWDDIISQASRNVEILKDPEAVKQLVSILKTNVRACRALAHPYVVQLGRIYLDMLNVYKVMSENISQAIALNGVAVTKQPLIKNMRIIKKETLNLISSWVTRSIDNSMVLENFIPPLLDAVLLDYQRTAVPEAREPEVLSCMAAIVHRLEGHITSEVPKIFDAVFECTLEMINKDFEEYPEHRTEFFLLLQAVNTNCFKAFLSIPPAQFKLVLDSIIWAFKHTMRNVADTGLQILYRLLLNVEEHPQAAQSFYRTYLCDILEHVFSVVTDTSHGAGLTMHATILAHIFSLVEAGRVTVPLGPTPDNVLYIQEYIANLLKTAFSHLTDNQIKITVQGLFNLNQDIPAFKDHLRDFLVQIREYTGEDDSDLYLEERLFVLRQAQEDKRTVQLSVPGILNPHELPEEMQD
Summary
Uniprot
H9JMA4
A0A1E1WF48
A0A2A4J8R0
A0A0S2MU13
A0A212EH89
A0A0N1PJF4
+ More
A0A194PNK6 S4PE76 A0A3S2L8T7 A0A0L7LSX2 A0A067R5K6 A0A151IP52 A0A0J7KFR1 A0A195FVZ2 A0A2J7R911 F4WNS6 A0A195B8P6 A0A158NW25 A0A195ECH5 A0A026WJN7 K7IQJ4 A0A232FE09 A0A154PI79 A0A0V0G423 A0A224XH56 A0A069DXT0 A0A087ZMS1 D6X0Q6 A0A0P4VTA0 A0A0T6AZF3 A0A1Y1MZ97 A0A2A3E6Y5 A0A0A9WM64 A0A0K8SDD3 A0A1B6DS12 A0A1B6JXF3 A0A151X7I5 A0A1W4XPR0 N6TJI2 A0A293MVZ7 A0A0P4WCE4 A0A0P4WHK7 A0A2R5L7Z6 A0A087TAY2 A0A1D2NK16 V5I1V1 A0A131YVA4 A0A1J1I615 A0A1W5A0P4 A0A3Q3S1N3 A0A3B3I741 A0A3P9LG22 L7M5Z0 A0A131XFJ1 A0A218URZ2 A0A093FS26 A0A3Q1EGB9 U3I8L1 A0A1D5P8H7 U3K8W1 A0A3P9CR46 A0A1U7RM64 A0A0F8B6D8 A0A3Q2W904 A0A315VUI3 A0A3P8PZM5 A0A091QE91 A0A1A7Y3Q1 A0A3N0XW18 A0A3B3B4I4 A0A1A8ELR3 A0A3B3VUG1 A0A250Y6Q4 D2HZX2 E2R9K4 A0A341C1V7 L8IXM5 G3TDG6 U6CR70 A0A3Q2I6B8 A0A2U4BZ49 A0A340XSA9 A0A2Y9EU68 A0A2Y9HZK7 M3VYZ8 A0A2Y9JG64 A0A3Q7RRM9 A0A3P4MAY6 A0A1U7TKR2 A0A2Y9QBU2 E1BE98 M3Y2G7 A0A2K6G794 A0A2U3VRC8 A0A3Q7VSE8 A0A3Q7NT77 A0A384AR15 A0A3B3UPB7 A0A3P9NZU7 A0A3B5R002
A0A194PNK6 S4PE76 A0A3S2L8T7 A0A0L7LSX2 A0A067R5K6 A0A151IP52 A0A0J7KFR1 A0A195FVZ2 A0A2J7R911 F4WNS6 A0A195B8P6 A0A158NW25 A0A195ECH5 A0A026WJN7 K7IQJ4 A0A232FE09 A0A154PI79 A0A0V0G423 A0A224XH56 A0A069DXT0 A0A087ZMS1 D6X0Q6 A0A0P4VTA0 A0A0T6AZF3 A0A1Y1MZ97 A0A2A3E6Y5 A0A0A9WM64 A0A0K8SDD3 A0A1B6DS12 A0A1B6JXF3 A0A151X7I5 A0A1W4XPR0 N6TJI2 A0A293MVZ7 A0A0P4WCE4 A0A0P4WHK7 A0A2R5L7Z6 A0A087TAY2 A0A1D2NK16 V5I1V1 A0A131YVA4 A0A1J1I615 A0A1W5A0P4 A0A3Q3S1N3 A0A3B3I741 A0A3P9LG22 L7M5Z0 A0A131XFJ1 A0A218URZ2 A0A093FS26 A0A3Q1EGB9 U3I8L1 A0A1D5P8H7 U3K8W1 A0A3P9CR46 A0A1U7RM64 A0A0F8B6D8 A0A3Q2W904 A0A315VUI3 A0A3P8PZM5 A0A091QE91 A0A1A7Y3Q1 A0A3N0XW18 A0A3B3B4I4 A0A1A8ELR3 A0A3B3VUG1 A0A250Y6Q4 D2HZX2 E2R9K4 A0A341C1V7 L8IXM5 G3TDG6 U6CR70 A0A3Q2I6B8 A0A2U4BZ49 A0A340XSA9 A0A2Y9EU68 A0A2Y9HZK7 M3VYZ8 A0A2Y9JG64 A0A3Q7RRM9 A0A3P4MAY6 A0A1U7TKR2 A0A2Y9QBU2 E1BE98 M3Y2G7 A0A2K6G794 A0A2U3VRC8 A0A3Q7VSE8 A0A3Q7NT77 A0A384AR15 A0A3B3UPB7 A0A3P9NZU7 A0A3B5R002
Pubmed
19121390
22118469
26354079
23622113
26227816
24845553
+ More
21719571 21347285 24508170 30249741 20075255 28648823 26334808 18362917 19820115 27129103 28004739 25401762 26823975 23537049 27289101 25765539 26830274 17554307 25576852 28049606 23749191 15592404 25186727 25835551 29703783 29451363 28087693 20010809 16341006 22751099 19892987 17975172 19393038 23542700
21719571 21347285 24508170 30249741 20075255 28648823 26334808 18362917 19820115 27129103 28004739 25401762 26823975 23537049 27289101 25765539 26830274 17554307 25576852 28049606 23749191 15592404 25186727 25835551 29703783 29451363 28087693 20010809 16341006 22751099 19892987 17975172 19393038 23542700
EMBL
BABH01033370
BABH01033371
GDQN01005430
JAT85624.1
NWSH01002640
PCG67783.1
+ More
KT208379 ODYU01002368 ALO79336.1 SOQ39780.1 AGBW02014932 OWR40846.1 KQ459752 KPJ20201.1 KQ459603 KPI92705.1 GAIX01003281 JAA89279.1 RSAL01000504 RVE41519.1 JTDY01000153 KOB78568.1 KK852870 KDR14611.1 KQ976892 KYN07249.1 LBMM01007957 KMQ89268.1 KQ981215 KYN44487.1 NEVH01006721 PNF37319.1 GL888237 EGI64269.1 KQ976565 KYM80564.1 ADTU01002913 KQ979074 KYN22811.1 KK107168 QOIP01000008 EZA56215.1 RLU19561.1 NNAY01000360 OXU28902.1 KQ434899 KZC10900.1 GECL01003306 JAP02818.1 GFTR01008514 JAW07912.1 GBGD01000253 JAC88636.1 KQ971371 EFA09995.1 GDKW01000762 JAI55833.1 LJIG01022446 KRT80563.1 GEZM01017018 JAV90964.1 KZ288363 PBC26929.1 GBHO01035093 GBHO01015441 GDHC01008944 GDHC01000287 JAG08511.1 JAG28163.1 JAQ09685.1 JAQ18342.1 GBRD01017708 GBRD01015026 JAG50800.1 GEDC01008855 JAS28443.1 GECU01004133 JAT03574.1 KQ982449 KYQ56280.1 APGK01035522 APGK01035523 KB740926 ENN78028.1 GFWV01018762 MAA43490.1 GDRN01052587 JAI66278.1 GDRN01052586 JAI66279.1 GGLE01001473 MBY05599.1 KK114363 KFM62271.1 LJIJ01000020 ODN05594.1 GANP01003030 JAB81438.1 GEDV01006147 JAP82410.1 CVRI01000038 CRK94382.1 GACK01005323 JAA59711.1 GEFH01003683 JAP64898.1 MUZQ01000154 OWK56533.1 KK400076 KFV59480.1 ADON01053384 ADON01053385 AADN05000009 AGTO01011165 KQ041519 KKF25848.1 NHOQ01001229 PWA26065.1 KK696691 KFQ24792.1 HADW01020362 HADX01002373 SBP24605.1 RJVU01059333 ROJ78905.1 HAEB01000849 SBQ47324.1 GFFV01000660 JAV39285.1 GL193845 EFB24524.1 AAEX03007556 JH880550 ELR60723.1 HAAF01001150 CCP72976.1 AANG04002548 CYRY02009931 VCW78129.1 AEYP01010098
KT208379 ODYU01002368 ALO79336.1 SOQ39780.1 AGBW02014932 OWR40846.1 KQ459752 KPJ20201.1 KQ459603 KPI92705.1 GAIX01003281 JAA89279.1 RSAL01000504 RVE41519.1 JTDY01000153 KOB78568.1 KK852870 KDR14611.1 KQ976892 KYN07249.1 LBMM01007957 KMQ89268.1 KQ981215 KYN44487.1 NEVH01006721 PNF37319.1 GL888237 EGI64269.1 KQ976565 KYM80564.1 ADTU01002913 KQ979074 KYN22811.1 KK107168 QOIP01000008 EZA56215.1 RLU19561.1 NNAY01000360 OXU28902.1 KQ434899 KZC10900.1 GECL01003306 JAP02818.1 GFTR01008514 JAW07912.1 GBGD01000253 JAC88636.1 KQ971371 EFA09995.1 GDKW01000762 JAI55833.1 LJIG01022446 KRT80563.1 GEZM01017018 JAV90964.1 KZ288363 PBC26929.1 GBHO01035093 GBHO01015441 GDHC01008944 GDHC01000287 JAG08511.1 JAG28163.1 JAQ09685.1 JAQ18342.1 GBRD01017708 GBRD01015026 JAG50800.1 GEDC01008855 JAS28443.1 GECU01004133 JAT03574.1 KQ982449 KYQ56280.1 APGK01035522 APGK01035523 KB740926 ENN78028.1 GFWV01018762 MAA43490.1 GDRN01052587 JAI66278.1 GDRN01052586 JAI66279.1 GGLE01001473 MBY05599.1 KK114363 KFM62271.1 LJIJ01000020 ODN05594.1 GANP01003030 JAB81438.1 GEDV01006147 JAP82410.1 CVRI01000038 CRK94382.1 GACK01005323 JAA59711.1 GEFH01003683 JAP64898.1 MUZQ01000154 OWK56533.1 KK400076 KFV59480.1 ADON01053384 ADON01053385 AADN05000009 AGTO01011165 KQ041519 KKF25848.1 NHOQ01001229 PWA26065.1 KK696691 KFQ24792.1 HADW01020362 HADX01002373 SBP24605.1 RJVU01059333 ROJ78905.1 HAEB01000849 SBQ47324.1 GFFV01000660 JAV39285.1 GL193845 EFB24524.1 AAEX03007556 JH880550 ELR60723.1 HAAF01001150 CCP72976.1 AANG04002548 CYRY02009931 VCW78129.1 AEYP01010098
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000037510 UP000027135 UP000078542 UP000036403 UP000078541 UP000235965 UP000007755 UP000078540 UP000005205 UP000078492 UP000053097 UP000279307 UP000002358 UP000215335 UP000076502 UP000005203 UP000007266 UP000242457 UP000075809 UP000192223 UP000019118 UP000054359 UP000094527 UP000183832 UP000192224 UP000261640 UP000001038 UP000265180 UP000197619 UP000257200 UP000016666 UP000000539 UP000016665 UP000265160 UP000189705 UP000264840 UP000265100 UP000261560 UP000261500 UP000002254 UP000252040 UP000007646 UP000002281 UP000245320 UP000265300 UP000248484 UP000248481 UP000011712 UP000248482 UP000286640 UP000189704 UP000248483 UP000009136 UP000000715 UP000233160 UP000245340 UP000286642 UP000286641 UP000261681 UP000242638 UP000002852
UP000037510 UP000027135 UP000078542 UP000036403 UP000078541 UP000235965 UP000007755 UP000078540 UP000005205 UP000078492 UP000053097 UP000279307 UP000002358 UP000215335 UP000076502 UP000005203 UP000007266 UP000242457 UP000075809 UP000192223 UP000019118 UP000054359 UP000094527 UP000183832 UP000192224 UP000261640 UP000001038 UP000265180 UP000197619 UP000257200 UP000016666 UP000000539 UP000016665 UP000265160 UP000189705 UP000264840 UP000265100 UP000261560 UP000261500 UP000002254 UP000252040 UP000007646 UP000002281 UP000245320 UP000265300 UP000248484 UP000248481 UP000011712 UP000248482 UP000286640 UP000189704 UP000248483 UP000009136 UP000000715 UP000233160 UP000245340 UP000286642 UP000286641 UP000261681 UP000242638 UP000002852
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JMA4
A0A1E1WF48
A0A2A4J8R0
A0A0S2MU13
A0A212EH89
A0A0N1PJF4
+ More
A0A194PNK6 S4PE76 A0A3S2L8T7 A0A0L7LSX2 A0A067R5K6 A0A151IP52 A0A0J7KFR1 A0A195FVZ2 A0A2J7R911 F4WNS6 A0A195B8P6 A0A158NW25 A0A195ECH5 A0A026WJN7 K7IQJ4 A0A232FE09 A0A154PI79 A0A0V0G423 A0A224XH56 A0A069DXT0 A0A087ZMS1 D6X0Q6 A0A0P4VTA0 A0A0T6AZF3 A0A1Y1MZ97 A0A2A3E6Y5 A0A0A9WM64 A0A0K8SDD3 A0A1B6DS12 A0A1B6JXF3 A0A151X7I5 A0A1W4XPR0 N6TJI2 A0A293MVZ7 A0A0P4WCE4 A0A0P4WHK7 A0A2R5L7Z6 A0A087TAY2 A0A1D2NK16 V5I1V1 A0A131YVA4 A0A1J1I615 A0A1W5A0P4 A0A3Q3S1N3 A0A3B3I741 A0A3P9LG22 L7M5Z0 A0A131XFJ1 A0A218URZ2 A0A093FS26 A0A3Q1EGB9 U3I8L1 A0A1D5P8H7 U3K8W1 A0A3P9CR46 A0A1U7RM64 A0A0F8B6D8 A0A3Q2W904 A0A315VUI3 A0A3P8PZM5 A0A091QE91 A0A1A7Y3Q1 A0A3N0XW18 A0A3B3B4I4 A0A1A8ELR3 A0A3B3VUG1 A0A250Y6Q4 D2HZX2 E2R9K4 A0A341C1V7 L8IXM5 G3TDG6 U6CR70 A0A3Q2I6B8 A0A2U4BZ49 A0A340XSA9 A0A2Y9EU68 A0A2Y9HZK7 M3VYZ8 A0A2Y9JG64 A0A3Q7RRM9 A0A3P4MAY6 A0A1U7TKR2 A0A2Y9QBU2 E1BE98 M3Y2G7 A0A2K6G794 A0A2U3VRC8 A0A3Q7VSE8 A0A3Q7NT77 A0A384AR15 A0A3B3UPB7 A0A3P9NZU7 A0A3B5R002
A0A194PNK6 S4PE76 A0A3S2L8T7 A0A0L7LSX2 A0A067R5K6 A0A151IP52 A0A0J7KFR1 A0A195FVZ2 A0A2J7R911 F4WNS6 A0A195B8P6 A0A158NW25 A0A195ECH5 A0A026WJN7 K7IQJ4 A0A232FE09 A0A154PI79 A0A0V0G423 A0A224XH56 A0A069DXT0 A0A087ZMS1 D6X0Q6 A0A0P4VTA0 A0A0T6AZF3 A0A1Y1MZ97 A0A2A3E6Y5 A0A0A9WM64 A0A0K8SDD3 A0A1B6DS12 A0A1B6JXF3 A0A151X7I5 A0A1W4XPR0 N6TJI2 A0A293MVZ7 A0A0P4WCE4 A0A0P4WHK7 A0A2R5L7Z6 A0A087TAY2 A0A1D2NK16 V5I1V1 A0A131YVA4 A0A1J1I615 A0A1W5A0P4 A0A3Q3S1N3 A0A3B3I741 A0A3P9LG22 L7M5Z0 A0A131XFJ1 A0A218URZ2 A0A093FS26 A0A3Q1EGB9 U3I8L1 A0A1D5P8H7 U3K8W1 A0A3P9CR46 A0A1U7RM64 A0A0F8B6D8 A0A3Q2W904 A0A315VUI3 A0A3P8PZM5 A0A091QE91 A0A1A7Y3Q1 A0A3N0XW18 A0A3B3B4I4 A0A1A8ELR3 A0A3B3VUG1 A0A250Y6Q4 D2HZX2 E2R9K4 A0A341C1V7 L8IXM5 G3TDG6 U6CR70 A0A3Q2I6B8 A0A2U4BZ49 A0A340XSA9 A0A2Y9EU68 A0A2Y9HZK7 M3VYZ8 A0A2Y9JG64 A0A3Q7RRM9 A0A3P4MAY6 A0A1U7TKR2 A0A2Y9QBU2 E1BE98 M3Y2G7 A0A2K6G794 A0A2U3VRC8 A0A3Q7VSE8 A0A3Q7NT77 A0A384AR15 A0A3B3UPB7 A0A3P9NZU7 A0A3B5R002
PDB
3NC1
E-value=0,
Score=4160
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
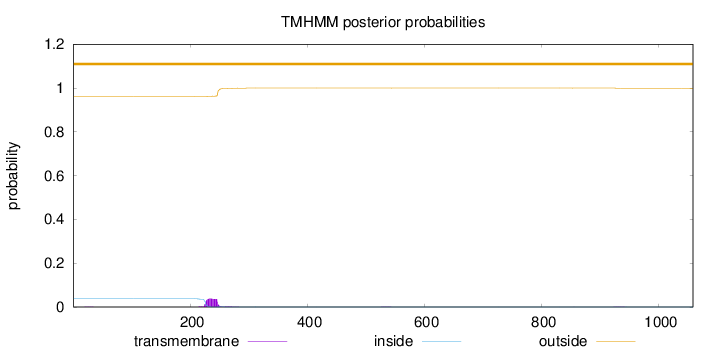
Length:
1058
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.877929999999997
Exp number, first 60 AAs:
0.00402
Total prob of N-in:
0.03759
outside
1 - 1058
Population Genetic Test Statistics
Pi
2.318408
Theta
3.604918
Tajima's D
0.941066
CLR
0.928852
CSRT
0.67951602419879
Interpretation
Uncertain
