Gene
KWMTBOMO07297
Pre Gene Modal
BGIBMGA010655
Annotation
PREDICTED:_menin_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.686 Nuclear Reliability : 1.383
Sequence
CDS
ATGAGTGGAATTAATGATTACAAACACTTTTTCCCTATTGACGGCATACAGAAAGTATGCGATTTATTTGAAAAACAGCTACGCAGTGACAAAGAGGCGGATTTAGCATTATTCTCCATTATTGTCGGTGCGGTTGAAAACAATTTAACAAATATTAAAATAGGTGATAAAGATGTGAATTTGCATTCAAATAATTTCGTTAAAATGAAATTTCCATGTATTGAATGGGATGAAGTGCGTTCGCTTCATGAACGATTTGTTAACCTCATGCGTACCGGCGTAGATGCAAAGCTCCTGAGTGGCGCTTACGCTACAAGAGATTTAGTTAAAAAGGTTGCGGACGTCGTCTGGAATTCATTAACTCGCAGTTATTACAAAGACCGGGCGCATTTGCAGTCTATTTACAGTTTCTTAACAGGCAATAAGTTAGATTGTTTTGGTGTTGCATTTGCTGTTACTGCTGGTTGTCAAGTATTGGGTAAACAAGACGTACATCTCGCTTTGTCTGAAGATCATGCCTGGGTAGTTTTTGGACAAGATGGAAAGGAAACGGCAGAAGTGACTTGGCATGGTAAAGGCAATGAAGACAAAAGAGGCAGGTCAGTCGGAGATGGTGTATCAGCTCGGTCGTGGTTGTACGTTGCAGGGAAACCAGTAGTATGCACAAGGGTAATGGAGTTGGCAGCACTGGTATCGTCAATTAATACAACTTTAACTCAAACTGTGGATGTGCATGAAGTAGCTCAAATGCAACATAAACTCCTCTGGTTACTTTATGATAGTGGATATTTGGATGCATACCCAATGGCATTGGGCAACTTAGCTGATCTAGATGATTATATGAAAGTACCAAATATAACAAAAGAAAATAATACAACCTCAAGTACTGATGATATCTCCAATGTACAAATGGACAATACACAGAGACCAGATGTAGCTGGGCTTTATGCTCAGGCTATTAAATCTGCCAAAACATATTATGATGATCGACATGTTTATCCTTACACACTCCAAGGAGGATATTATCACCGCCAAAAGATGTATAAAGAAGCGTTCCATGCATGGGCATCAGCTGGTGACGTTATTAGGCACTATAACTATTCTCGTGACGATGAAGAAATCTACAAGGAATTTGCAGAAATTGCTAATGAACTAATACCACAGTTAATGAAAGCTGAAAGTTCAGGACATTCAGCAAGGTCTATACTGAAGGATCCTGAATGTTTTGCATCACTATTAAGATTTTATGATGGAATCTGTAAATGGGAGGAAGGTAGTCAGACTCCAATACTGCACATAGGTTGGGCAAAGCCGCTTGTGAGCACTATATCGAAGTTTGATGCTGAAGTTAGGGCTCAAGTGTACATAGTATGTCGACCTGATTCTGGTGAAGTTCTCGAACACCAAGAACCGGACGGAGAAAAAAAAGAAACCACAGAAAACGGCAATGAATTAAAAAGTAACAATTCAGAAAACGCAGAAATGACTGTACGTGAACAATTAACGGCCGCTGTGAACAGTCGACCGCGTGTCACGTTATATAGTCATAAAATGGCTGCATTAAAGCCATTGCTGTTAGCAGAAAGACTCAATACGCATGCGTTACAATTACAACTAACAGCTCAAAGTCAGTTACAAAGACCTCAGGTACCAACTAAAAGACGAGACGATGAAGATCAAAATGCTTGTTTCCCGACAGCGCGGGCTAAGAGGGCACGACGTGAACGTTAA
Protein
MSGINDYKHFFPIDGIQKVCDLFEKQLRSDKEADLALFSIIVGAVENNLTNIKIGDKDVNLHSNNFVKMKFPCIEWDEVRSLHERFVNLMRTGVDAKLLSGAYATRDLVKKVADVVWNSLTRSYYKDRAHLQSIYSFLTGNKLDCFGVAFAVTAGCQVLGKQDVHLALSEDHAWVVFGQDGKETAEVTWHGKGNEDKRGRSVGDGVSARSWLYVAGKPVVCTRVMELAALVSSINTTLTQTVDVHEVAQMQHKLLWLLYDSGYLDAYPMALGNLADLDDYMKVPNITKENNTTSSTDDISNVQMDNTQRPDVAGLYAQAIKSAKTYYDDRHVYPYTLQGGYYHRQKMYKEAFHAWASAGDVIRHYNYSRDDEEIYKEFAEIANELIPQLMKAESSGHSARSILKDPECFASLLRFYDGICKWEEGSQTPILHIGWAKPLVSTISKFDAEVRAQVYIVCRPDSGEVLEHQEPDGEKKETTENGNELKSNNSENAEMTVREQLTAAVNSRPRVTLYSHKMAALKPLLLAERLNTHALQLQLTAQSQLQRPQVPTKRRDDEDQNACFPTARAKRARRER
Summary
Uniprot
H9JMA2
A0A2A4K231
A0A3S2NRZ9
A0A194PI29
A0A0L7LTK1
A0A212EH87
+ More
A0A2H1WFN7 A0A0N0PEM1 A0A067RWC5 A0A0T6B4P6 A0A1Y1NAX0 A0A2P8YIN8 A0A0P5HN77 A0A0N8AWJ6 A0A0P6DFV8 A0A0P5RZY2 A0A0P5SK52 A0A0P5DAD9 E9HQV9 D2A2K0 A0A0P5KMA5 A0A1B6DHZ8 A0A347ZJ85 A0A0P5SL01 A0A0P6IS43 A0A0P5EUP3 A0A0P5N1Y1 A0A0P5PJH2 A0A0P5HX16 A0A0P6FBP2 A0A0N8DKE8 A0A0P5DG25 A0A0N8A7E9 A0A0P5AJS5 A0A0P5CPJ2 A0A0P5CMZ3 A0A1W4W6V3 A0A1B6H5M7 A0A1B6KSE4 A0A0P5HVN1 E0VP28 T1HZF3 A0A0N8EE58 A0A023F2X2 A0A0P5DDE6 A0A0P5BM40 A0A0P5BIR0 A0A0P5FKE5 A0A0P4ZRE8 A0A3Q0IT98 U4ULM9 N6TRY1 A0A0A9WKM6 A0A310SL13 A0A154PD09 A0A0P6CWQ0 A0A151WQ51 E2A9C8 A0A0J7L9E2 A0A195CW09 A0A182GNU2 A0A0P4WMT2 A0A182F4W1 Q179C2 A0A2M4A7Z2 W5J4M2 A0A0M8ZYF2 A0A2M3Z431 A0A2M3Z439 A0A182PPJ6 A0A087ZYU0 A0A2M4BF74 A0A182RCU6 A0A2M4BF62 A0A232F8N6 E2C0Y0 A0A0L7RDW0 A0A2A3EFN1 A0A182QZA3 A0A182W724 A0A182T9T3 A0A182MMY8 A0A195F5P8 U5EUV1 E9IUU8 A0A084W886 A0A182IN88 A0A195AYP7 A0A158NYI8 A0A182L1F8 A0A182WU19 A0A182TXU0 A0A182HUN2 A0A026WQC4 A0A1S4GXD4 A0A182NUE7 A0A182VGH9 Q7PND1 A0A151J3Z2
A0A2H1WFN7 A0A0N0PEM1 A0A067RWC5 A0A0T6B4P6 A0A1Y1NAX0 A0A2P8YIN8 A0A0P5HN77 A0A0N8AWJ6 A0A0P6DFV8 A0A0P5RZY2 A0A0P5SK52 A0A0P5DAD9 E9HQV9 D2A2K0 A0A0P5KMA5 A0A1B6DHZ8 A0A347ZJ85 A0A0P5SL01 A0A0P6IS43 A0A0P5EUP3 A0A0P5N1Y1 A0A0P5PJH2 A0A0P5HX16 A0A0P6FBP2 A0A0N8DKE8 A0A0P5DG25 A0A0N8A7E9 A0A0P5AJS5 A0A0P5CPJ2 A0A0P5CMZ3 A0A1W4W6V3 A0A1B6H5M7 A0A1B6KSE4 A0A0P5HVN1 E0VP28 T1HZF3 A0A0N8EE58 A0A023F2X2 A0A0P5DDE6 A0A0P5BM40 A0A0P5BIR0 A0A0P5FKE5 A0A0P4ZRE8 A0A3Q0IT98 U4ULM9 N6TRY1 A0A0A9WKM6 A0A310SL13 A0A154PD09 A0A0P6CWQ0 A0A151WQ51 E2A9C8 A0A0J7L9E2 A0A195CW09 A0A182GNU2 A0A0P4WMT2 A0A182F4W1 Q179C2 A0A2M4A7Z2 W5J4M2 A0A0M8ZYF2 A0A2M3Z431 A0A2M3Z439 A0A182PPJ6 A0A087ZYU0 A0A2M4BF74 A0A182RCU6 A0A2M4BF62 A0A232F8N6 E2C0Y0 A0A0L7RDW0 A0A2A3EFN1 A0A182QZA3 A0A182W724 A0A182T9T3 A0A182MMY8 A0A195F5P8 U5EUV1 E9IUU8 A0A084W886 A0A182IN88 A0A195AYP7 A0A158NYI8 A0A182L1F8 A0A182WU19 A0A182TXU0 A0A182HUN2 A0A026WQC4 A0A1S4GXD4 A0A182NUE7 A0A182VGH9 Q7PND1 A0A151J3Z2
Pubmed
EMBL
BABH01033367
BABH01033368
NWSH01000241
PCG78076.1
RSAL01000282
RVE43029.1
+ More
KQ459603 KPI92708.1 JTDY01000153 KOB78566.1 AGBW02014932 OWR40849.1 ODYU01008353 SOQ51871.1 KQ459752 KPJ20199.1 KK852424 KDR24179.1 LJIG01009798 KRT82360.1 GEZM01007610 JAV95062.1 PYGN01000568 PSN44102.1 GDIQ01224941 JAK26784.1 GDIQ01235891 JAK15834.1 GDIQ01078237 JAN16500.1 GDIQ01103277 JAL48449.1 GDIQ01103276 JAL48450.1 GDIP01175146 JAJ48256.1 GL732727 EFX65871.1 KQ971338 EFA02211.1 GDIQ01183261 JAK68464.1 GEDC01025299 GEDC01012026 JAS11999.1 JAS25272.1 FX985653 BBA84391.1 GDIP01142029 JAL61685.1 GDIQ01000978 JAN93759.1 GDIQ01267016 JAJ84708.1 GDIQ01170265 JAK81460.1 GDIQ01132732 JAL18994.1 GDIQ01222346 JAK29379.1 GDIQ01065858 JAN28879.1 GDIP01025063 JAM78652.1 GDIP01157982 JAJ65420.1 GDIP01175145 LRGB01000243 JAJ48257.1 KZS20236.1 GDIP01211580 JAJ11822.1 GDIP01167279 JAJ56123.1 GDIP01168181 JAJ55221.1 GECU01037802 JAS69904.1 GEBQ01025618 JAT14359.1 GDIQ01227239 JAK24486.1 DS235354 EEB15134.1 ACPB03008638 GDIQ01035604 JAN59133.1 GBBI01003017 JAC15695.1 GDIP01157981 JAJ65421.1 GDIP01182735 JAJ40667.1 GDIP01184181 JAJ39221.1 GDIQ01254127 JAJ97597.1 GDIP01213389 JAJ10013.1 KB632354 ERL93368.1 APGK01057288 KB741279 ENN71051.1 GBHO01036563 GBHO01036561 GBHO01033101 GBHO01021250 GDHC01016611 GDHC01012970 GDHC01004058 JAG07041.1 JAG07043.1 JAG10503.1 JAG22354.1 JAQ02018.1 JAQ05659.1 JAQ14571.1 KQ761768 OAD57069.1 KQ434864 KZC09090.1 GDIQ01086354 JAN08383.1 KQ982843 KYQ50032.1 GL437781 EFN69970.1 LBMM01000214 KMR04463.1 KQ977220 KYN04868.1 JXUM01077114 JXUM01077115 KQ562988 KXJ74699.1 GDRN01003952 JAI67986.1 CH477355 EAT42797.1 GGFK01003437 MBW36758.1 ADMH02002111 ETN58871.1 KQ435824 KOX72153.1 GGFM01002531 MBW23282.1 GGFM01002540 MBW23291.1 GGFJ01002553 MBW51694.1 GGFJ01002548 MBW51689.1 NNAY01000715 OXU26838.1 GL451850 EFN78416.1 KQ414613 KOC68990.1 KZ288269 PBC29966.1 AXCN02000762 AXCM01001263 KQ981768 KYN35915.1 GANO01003702 JAB56169.1 GL766050 EFZ15660.1 ATLV01021388 KE525317 KFB46430.1 KQ976701 KYM77321.1 ADTU01003927 APCN01002467 KK107151 QOIP01000003 EZA57314.1 RLU24426.1 AAAB01008964 EAA12374.5 KQ980228 KYN17148.1
KQ459603 KPI92708.1 JTDY01000153 KOB78566.1 AGBW02014932 OWR40849.1 ODYU01008353 SOQ51871.1 KQ459752 KPJ20199.1 KK852424 KDR24179.1 LJIG01009798 KRT82360.1 GEZM01007610 JAV95062.1 PYGN01000568 PSN44102.1 GDIQ01224941 JAK26784.1 GDIQ01235891 JAK15834.1 GDIQ01078237 JAN16500.1 GDIQ01103277 JAL48449.1 GDIQ01103276 JAL48450.1 GDIP01175146 JAJ48256.1 GL732727 EFX65871.1 KQ971338 EFA02211.1 GDIQ01183261 JAK68464.1 GEDC01025299 GEDC01012026 JAS11999.1 JAS25272.1 FX985653 BBA84391.1 GDIP01142029 JAL61685.1 GDIQ01000978 JAN93759.1 GDIQ01267016 JAJ84708.1 GDIQ01170265 JAK81460.1 GDIQ01132732 JAL18994.1 GDIQ01222346 JAK29379.1 GDIQ01065858 JAN28879.1 GDIP01025063 JAM78652.1 GDIP01157982 JAJ65420.1 GDIP01175145 LRGB01000243 JAJ48257.1 KZS20236.1 GDIP01211580 JAJ11822.1 GDIP01167279 JAJ56123.1 GDIP01168181 JAJ55221.1 GECU01037802 JAS69904.1 GEBQ01025618 JAT14359.1 GDIQ01227239 JAK24486.1 DS235354 EEB15134.1 ACPB03008638 GDIQ01035604 JAN59133.1 GBBI01003017 JAC15695.1 GDIP01157981 JAJ65421.1 GDIP01182735 JAJ40667.1 GDIP01184181 JAJ39221.1 GDIQ01254127 JAJ97597.1 GDIP01213389 JAJ10013.1 KB632354 ERL93368.1 APGK01057288 KB741279 ENN71051.1 GBHO01036563 GBHO01036561 GBHO01033101 GBHO01021250 GDHC01016611 GDHC01012970 GDHC01004058 JAG07041.1 JAG07043.1 JAG10503.1 JAG22354.1 JAQ02018.1 JAQ05659.1 JAQ14571.1 KQ761768 OAD57069.1 KQ434864 KZC09090.1 GDIQ01086354 JAN08383.1 KQ982843 KYQ50032.1 GL437781 EFN69970.1 LBMM01000214 KMR04463.1 KQ977220 KYN04868.1 JXUM01077114 JXUM01077115 KQ562988 KXJ74699.1 GDRN01003952 JAI67986.1 CH477355 EAT42797.1 GGFK01003437 MBW36758.1 ADMH02002111 ETN58871.1 KQ435824 KOX72153.1 GGFM01002531 MBW23282.1 GGFM01002540 MBW23291.1 GGFJ01002553 MBW51694.1 GGFJ01002548 MBW51689.1 NNAY01000715 OXU26838.1 GL451850 EFN78416.1 KQ414613 KOC68990.1 KZ288269 PBC29966.1 AXCN02000762 AXCM01001263 KQ981768 KYN35915.1 GANO01003702 JAB56169.1 GL766050 EFZ15660.1 ATLV01021388 KE525317 KFB46430.1 KQ976701 KYM77321.1 ADTU01003927 APCN01002467 KK107151 QOIP01000003 EZA57314.1 RLU24426.1 AAAB01008964 EAA12374.5 KQ980228 KYN17148.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000037510
UP000007151
+ More
UP000053240 UP000027135 UP000245037 UP000000305 UP000007266 UP000076858 UP000192223 UP000009046 UP000015103 UP000079169 UP000030742 UP000019118 UP000076502 UP000075809 UP000000311 UP000036403 UP000078542 UP000069940 UP000249989 UP000069272 UP000008820 UP000000673 UP000053105 UP000075885 UP000005203 UP000075900 UP000215335 UP000008237 UP000053825 UP000242457 UP000075886 UP000075920 UP000075901 UP000075883 UP000078541 UP000030765 UP000075880 UP000078540 UP000005205 UP000075882 UP000076407 UP000075902 UP000075840 UP000053097 UP000279307 UP000075884 UP000075903 UP000007062 UP000078492
UP000053240 UP000027135 UP000245037 UP000000305 UP000007266 UP000076858 UP000192223 UP000009046 UP000015103 UP000079169 UP000030742 UP000019118 UP000076502 UP000075809 UP000000311 UP000036403 UP000078542 UP000069940 UP000249989 UP000069272 UP000008820 UP000000673 UP000053105 UP000075885 UP000005203 UP000075900 UP000215335 UP000008237 UP000053825 UP000242457 UP000075886 UP000075920 UP000075901 UP000075883 UP000078541 UP000030765 UP000075880 UP000078540 UP000005205 UP000075882 UP000076407 UP000075902 UP000075840 UP000053097 UP000279307 UP000075884 UP000075903 UP000007062 UP000078492
PRIDE
Pfam
PF05053 Menin
Interpro
IPR007747
Menin
CDD
ProteinModelPortal
H9JMA2
A0A2A4K231
A0A3S2NRZ9
A0A194PI29
A0A0L7LTK1
A0A212EH87
+ More
A0A2H1WFN7 A0A0N0PEM1 A0A067RWC5 A0A0T6B4P6 A0A1Y1NAX0 A0A2P8YIN8 A0A0P5HN77 A0A0N8AWJ6 A0A0P6DFV8 A0A0P5RZY2 A0A0P5SK52 A0A0P5DAD9 E9HQV9 D2A2K0 A0A0P5KMA5 A0A1B6DHZ8 A0A347ZJ85 A0A0P5SL01 A0A0P6IS43 A0A0P5EUP3 A0A0P5N1Y1 A0A0P5PJH2 A0A0P5HX16 A0A0P6FBP2 A0A0N8DKE8 A0A0P5DG25 A0A0N8A7E9 A0A0P5AJS5 A0A0P5CPJ2 A0A0P5CMZ3 A0A1W4W6V3 A0A1B6H5M7 A0A1B6KSE4 A0A0P5HVN1 E0VP28 T1HZF3 A0A0N8EE58 A0A023F2X2 A0A0P5DDE6 A0A0P5BM40 A0A0P5BIR0 A0A0P5FKE5 A0A0P4ZRE8 A0A3Q0IT98 U4ULM9 N6TRY1 A0A0A9WKM6 A0A310SL13 A0A154PD09 A0A0P6CWQ0 A0A151WQ51 E2A9C8 A0A0J7L9E2 A0A195CW09 A0A182GNU2 A0A0P4WMT2 A0A182F4W1 Q179C2 A0A2M4A7Z2 W5J4M2 A0A0M8ZYF2 A0A2M3Z431 A0A2M3Z439 A0A182PPJ6 A0A087ZYU0 A0A2M4BF74 A0A182RCU6 A0A2M4BF62 A0A232F8N6 E2C0Y0 A0A0L7RDW0 A0A2A3EFN1 A0A182QZA3 A0A182W724 A0A182T9T3 A0A182MMY8 A0A195F5P8 U5EUV1 E9IUU8 A0A084W886 A0A182IN88 A0A195AYP7 A0A158NYI8 A0A182L1F8 A0A182WU19 A0A182TXU0 A0A182HUN2 A0A026WQC4 A0A1S4GXD4 A0A182NUE7 A0A182VGH9 Q7PND1 A0A151J3Z2
A0A2H1WFN7 A0A0N0PEM1 A0A067RWC5 A0A0T6B4P6 A0A1Y1NAX0 A0A2P8YIN8 A0A0P5HN77 A0A0N8AWJ6 A0A0P6DFV8 A0A0P5RZY2 A0A0P5SK52 A0A0P5DAD9 E9HQV9 D2A2K0 A0A0P5KMA5 A0A1B6DHZ8 A0A347ZJ85 A0A0P5SL01 A0A0P6IS43 A0A0P5EUP3 A0A0P5N1Y1 A0A0P5PJH2 A0A0P5HX16 A0A0P6FBP2 A0A0N8DKE8 A0A0P5DG25 A0A0N8A7E9 A0A0P5AJS5 A0A0P5CPJ2 A0A0P5CMZ3 A0A1W4W6V3 A0A1B6H5M7 A0A1B6KSE4 A0A0P5HVN1 E0VP28 T1HZF3 A0A0N8EE58 A0A023F2X2 A0A0P5DDE6 A0A0P5BM40 A0A0P5BIR0 A0A0P5FKE5 A0A0P4ZRE8 A0A3Q0IT98 U4ULM9 N6TRY1 A0A0A9WKM6 A0A310SL13 A0A154PD09 A0A0P6CWQ0 A0A151WQ51 E2A9C8 A0A0J7L9E2 A0A195CW09 A0A182GNU2 A0A0P4WMT2 A0A182F4W1 Q179C2 A0A2M4A7Z2 W5J4M2 A0A0M8ZYF2 A0A2M3Z431 A0A2M3Z439 A0A182PPJ6 A0A087ZYU0 A0A2M4BF74 A0A182RCU6 A0A2M4BF62 A0A232F8N6 E2C0Y0 A0A0L7RDW0 A0A2A3EFN1 A0A182QZA3 A0A182W724 A0A182T9T3 A0A182MMY8 A0A195F5P8 U5EUV1 E9IUU8 A0A084W886 A0A182IN88 A0A195AYP7 A0A158NYI8 A0A182L1F8 A0A182WU19 A0A182TXU0 A0A182HUN2 A0A026WQC4 A0A1S4GXD4 A0A182NUE7 A0A182VGH9 Q7PND1 A0A151J3Z2
PDB
4OG5
E-value=4.77855e-121,
Score=1114
Ontologies
GO
PANTHER
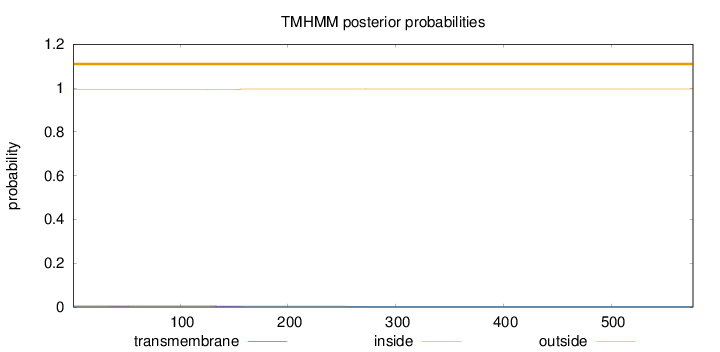
Topology
Length:
576
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0915599999999999
Exp number, first 60 AAs:
0.01054
Total prob of N-in:
0.00781
outside
1 - 576
Population Genetic Test Statistics
Pi
17.065913
Theta
15.688747
Tajima's D
0.799778
CLR
1.397495
CSRT
0.615419229038548
Interpretation
Uncertain
