Pre Gene Modal
BGIBMGA010653
Annotation
PREDICTED:_uncharacterized_protein_LOC106106714_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.149
Sequence
CDS
ATGCTTCGAAATAATTTAAAAAATTTGATAAAAAGTTGTCGTAGTGTACAACTTCAAATAATAGATTCTGTTGGGACAAATCATATAATAACAAATCAAAGAAGATTTAAAAGTGACCTATATGAACCTGATTACCTGATTAGCTTACAACCTGATGAGCCAGTCTATGACTGTTTGAACTTGCAACTTAAAGGTTATGATTATGCTATATTGGAAGCGTGCCAAAGTCAGATCCATAGATATGCTGAAGTGATGGGATTACAAGTAGAGGACTGTTGGGCGACCCCTGCTCAGCAGTTAAAGGTACAACGGTTCAAGCCAGGTAGTACTGCTCTAGATGCTGAATACCAATTGAATATATATGAGCGCAATGTTCAGGTTGTTGATGTTCCTGCATGGGCTTTGGGTACTCTGCTTCGGGTATCAAGGGCTCTTTTACCGGAGGGATGCACTTTGAATGTTCATGAACACACTCCTGAACATGAAGAGATTAGATATGTGCCAGACAACCAACTTATTGATCTGAAACAGCAACTAGAAGGCATGGGTGGCAGTCGTGCCGATAAAAAGAAAAAGAGATAA
Protein
MLRNNLKNLIKSCRSVQLQIIDSVGTNHIITNQRRFKSDLYEPDYLISLQPDEPVYDCLNLQLKGYDYAILEACQSQIHRYAEVMGLQVEDCWATPAQQLKVQRFKPGSTALDAEYQLNIYERNVQVVDVPAWALGTLLRVSRALLPEGCTLNVHEHTPEHEEIRYVPDNQLIDLKQQLEGMGGSRADKKKKR
Summary
Uniprot
H9JMA0
A0A2H1VZ67
A0A2A4K2S0
A0A194PHC7
A0A212EJD1
A0A0N0PEP3
+ More
I4DQ32 A0A0K8TT15 Q16W93 B4LTU9 A0A1B6LJ98 A0A2M3ZBW7 B4MW66 A0A2M4C0T0 A0A2M4C0V2 A0A023EGR2 A0A1A9ZIU1 A0A0L0C6Y4 A0A0M4DZT5 A0A1Q3FB65 A0A2S2NU69 A0A2M4AXR5 A0A2M4AXT8 A0A1W4VE14 B4KGC6 A0A2M4AXU6 A0A2M4AY26 A0A2S2QIK0 W5JDT0 B3MUA2 A0A1B6F3D4 B4P3K6 B4JAR7 A0A1A9VXL5 A0A1A9XDS5 A0A1B0B4M6 A0A034VH23 A0A3B0KB71 A0A1A9W1R4 C4WU99 B3N8W8 A0A1L8ECZ6 Q29MS9 D3TRG7 B4G8S6 A0A1L8E130 A0A1L8E106 A0A182PQC8 T1E8Y6 A0A2H8TM75 A0A182VCY0 A0A1I8MEE8 A0A1I8QCC3 A0A0K8VT04 A0A182KRE7 A0A182X3N7 A7URB7 A0A182I3D1 A0A182INU7 A0A182K442 Q9VQ35 B4IDB1 W8B2K8 A0A1Y1MN04 A0A2R7VX26 A0A0J9QTY8 A0A182QKH5 A0A182HBE7 A0A182U116 E9GLK0 A0A336MCA6 A0A1D2N342 A0A3R7PZX4 A0A182M046 A0A162CC62 A0A1B0GKG9 A0A0N8EJK3 A0A087T145 A0A0P5HU45 A0A084WND1 A0A226DAZ2 K7IUM6 B0WNR2 U4UPU0 A0A182FF66 A0A232FCT4 A0A182T7F3 A0A0P5HIN0 A0A0P5Q436 D2A319 A0A195BVM9 A0A182NHT2 A0A2J7RCX3 A0A195CPS4 A0A023G7C7 G3MS65 A0A023G4N8 A0A023FGC5 A0A1J1I9P5 E2B3D1
I4DQ32 A0A0K8TT15 Q16W93 B4LTU9 A0A1B6LJ98 A0A2M3ZBW7 B4MW66 A0A2M4C0T0 A0A2M4C0V2 A0A023EGR2 A0A1A9ZIU1 A0A0L0C6Y4 A0A0M4DZT5 A0A1Q3FB65 A0A2S2NU69 A0A2M4AXR5 A0A2M4AXT8 A0A1W4VE14 B4KGC6 A0A2M4AXU6 A0A2M4AY26 A0A2S2QIK0 W5JDT0 B3MUA2 A0A1B6F3D4 B4P3K6 B4JAR7 A0A1A9VXL5 A0A1A9XDS5 A0A1B0B4M6 A0A034VH23 A0A3B0KB71 A0A1A9W1R4 C4WU99 B3N8W8 A0A1L8ECZ6 Q29MS9 D3TRG7 B4G8S6 A0A1L8E130 A0A1L8E106 A0A182PQC8 T1E8Y6 A0A2H8TM75 A0A182VCY0 A0A1I8MEE8 A0A1I8QCC3 A0A0K8VT04 A0A182KRE7 A0A182X3N7 A7URB7 A0A182I3D1 A0A182INU7 A0A182K442 Q9VQ35 B4IDB1 W8B2K8 A0A1Y1MN04 A0A2R7VX26 A0A0J9QTY8 A0A182QKH5 A0A182HBE7 A0A182U116 E9GLK0 A0A336MCA6 A0A1D2N342 A0A3R7PZX4 A0A182M046 A0A162CC62 A0A1B0GKG9 A0A0N8EJK3 A0A087T145 A0A0P5HU45 A0A084WND1 A0A226DAZ2 K7IUM6 B0WNR2 U4UPU0 A0A182FF66 A0A232FCT4 A0A182T7F3 A0A0P5HIN0 A0A0P5Q436 D2A319 A0A195BVM9 A0A182NHT2 A0A2J7RCX3 A0A195CPS4 A0A023G7C7 G3MS65 A0A023G4N8 A0A023FGC5 A0A1J1I9P5 E2B3D1
Pubmed
19121390
26354079
22118469
22651552
26369729
17510324
+ More
17994087 24945155 26108605 20920257 23761445 17550304 25348373 15632085 20353571 25315136 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 28004739 22936249 26483478 21292972 27289101 24438588 20075255 23537049 28648823 18362917 19820115 22216098 20798317
17994087 24945155 26108605 20920257 23761445 17550304 25348373 15632085 20353571 25315136 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 28004739 22936249 26483478 21292972 27289101 24438588 20075255 23537049 28648823 18362917 19820115 22216098 20798317
EMBL
BABH01033361
BABH01033362
ODYU01005347
SOQ46097.1
NWSH01000241
PCG78073.1
+ More
KQ459603 KPI92712.1 AGBW02014491 OWR41585.1 KQ459752 KPJ20196.1 AK403906 BAM20022.1 GDAI01000533 JAI17070.1 CH477572 EAT38858.1 CH940649 EDW64000.1 GEBQ01016222 JAT23755.1 GGFM01005293 MBW26044.1 CH963857 EDW75936.1 GGFJ01009795 MBW58936.1 GGFJ01009796 MBW58937.1 GAPW01005075 JAC08523.1 JRES01000827 KNC28030.1 CP012523 ALC38178.1 GFDL01010229 JAV24816.1 GGMR01008100 MBY20719.1 GGFK01012278 MBW45599.1 GGFK01012260 MBW45581.1 CH933807 EDW11113.1 GGFK01012279 MBW45600.1 GGFK01012385 MBW45706.1 GGMS01008354 MBY77557.1 ADMH02001806 ETN61050.1 CH902624 EDV33431.1 GECZ01025396 JAS44373.1 CM000157 EDW87273.1 CH916368 EDW03875.1 JXJN01008372 GAKP01017535 GAKP01017534 JAC41417.1 OUUW01000006 SPP82301.1 AK340976 BAH71469.1 CH954177 EDV57368.1 GFDG01002294 JAV16505.1 CH379060 EAL33614.2 CCAG010010432 EZ424019 ADD20295.1 CH479180 EDW28756.1 GFDF01001677 JAV12407.1 GFDF01001676 JAV12408.1 GAMD01001787 JAA99803.1 GFXV01003334 MBW15139.1 GDHF01010336 JAI41978.1 AAAB01008807 EDO64559.1 APCN01000205 AE014134 AY069759 AAF51347.1 AAL39904.1 AGB92415.1 CH480829 EDW45537.1 GAMC01011191 GAMC01011188 GAMC01011185 JAB95370.1 GEZM01026310 JAV87051.1 KK854135 PTY11889.1 CM002910 KMY87506.1 AXCN02000377 JXUM01031874 KQ560938 KXJ80266.1 GL732551 EFX79524.1 UFQT01000912 SSX27964.1 LJIJ01000262 ODM99696.1 QCYY01000826 ROT82415.1 AXCM01000691 LRGB01001005 KZS14012.1 AJWK01028865 GDIQ01020444 JAN74293.1 KK112918 KFM58834.1 GDIQ01228071 JAK23654.1 ATLV01024587 KE525352 KFB51725.1 LNIX01000025 OXA42712.1 AAZX01006018 DS232014 EDS31903.1 KB632390 ERL94513.1 NNAY01000458 OXU28259.1 GDIQ01232463 JAK19262.1 GDIQ01122154 JAL29572.1 KQ971338 EFA01967.1 KQ976401 KYM92632.1 NEVH01005883 PNF38672.1 KQ977444 KYN02743.1 GBBM01006638 JAC28780.1 JO844716 AEO36333.1 GBBM01006639 GBBM01006637 JAC28779.1 GBBK01003900 JAC20582.1 CVRI01000045 CRK96968.1 GL445323 EFN89809.1
KQ459603 KPI92712.1 AGBW02014491 OWR41585.1 KQ459752 KPJ20196.1 AK403906 BAM20022.1 GDAI01000533 JAI17070.1 CH477572 EAT38858.1 CH940649 EDW64000.1 GEBQ01016222 JAT23755.1 GGFM01005293 MBW26044.1 CH963857 EDW75936.1 GGFJ01009795 MBW58936.1 GGFJ01009796 MBW58937.1 GAPW01005075 JAC08523.1 JRES01000827 KNC28030.1 CP012523 ALC38178.1 GFDL01010229 JAV24816.1 GGMR01008100 MBY20719.1 GGFK01012278 MBW45599.1 GGFK01012260 MBW45581.1 CH933807 EDW11113.1 GGFK01012279 MBW45600.1 GGFK01012385 MBW45706.1 GGMS01008354 MBY77557.1 ADMH02001806 ETN61050.1 CH902624 EDV33431.1 GECZ01025396 JAS44373.1 CM000157 EDW87273.1 CH916368 EDW03875.1 JXJN01008372 GAKP01017535 GAKP01017534 JAC41417.1 OUUW01000006 SPP82301.1 AK340976 BAH71469.1 CH954177 EDV57368.1 GFDG01002294 JAV16505.1 CH379060 EAL33614.2 CCAG010010432 EZ424019 ADD20295.1 CH479180 EDW28756.1 GFDF01001677 JAV12407.1 GFDF01001676 JAV12408.1 GAMD01001787 JAA99803.1 GFXV01003334 MBW15139.1 GDHF01010336 JAI41978.1 AAAB01008807 EDO64559.1 APCN01000205 AE014134 AY069759 AAF51347.1 AAL39904.1 AGB92415.1 CH480829 EDW45537.1 GAMC01011191 GAMC01011188 GAMC01011185 JAB95370.1 GEZM01026310 JAV87051.1 KK854135 PTY11889.1 CM002910 KMY87506.1 AXCN02000377 JXUM01031874 KQ560938 KXJ80266.1 GL732551 EFX79524.1 UFQT01000912 SSX27964.1 LJIJ01000262 ODM99696.1 QCYY01000826 ROT82415.1 AXCM01000691 LRGB01001005 KZS14012.1 AJWK01028865 GDIQ01020444 JAN74293.1 KK112918 KFM58834.1 GDIQ01228071 JAK23654.1 ATLV01024587 KE525352 KFB51725.1 LNIX01000025 OXA42712.1 AAZX01006018 DS232014 EDS31903.1 KB632390 ERL94513.1 NNAY01000458 OXU28259.1 GDIQ01232463 JAK19262.1 GDIQ01122154 JAL29572.1 KQ971338 EFA01967.1 KQ976401 KYM92632.1 NEVH01005883 PNF38672.1 KQ977444 KYN02743.1 GBBM01006638 JAC28780.1 JO844716 AEO36333.1 GBBM01006639 GBBM01006637 JAC28779.1 GBBK01003900 JAC20582.1 CVRI01000045 CRK96968.1 GL445323 EFN89809.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000008820
+ More
UP000008792 UP000007798 UP000092445 UP000037069 UP000092553 UP000192221 UP000009192 UP000000673 UP000007801 UP000002282 UP000001070 UP000078200 UP000092443 UP000092460 UP000268350 UP000091820 UP000008711 UP000001819 UP000092444 UP000008744 UP000075885 UP000075903 UP000095301 UP000095300 UP000075882 UP000076407 UP000007062 UP000075840 UP000075880 UP000075881 UP000000803 UP000001292 UP000075886 UP000069940 UP000249989 UP000075902 UP000000305 UP000094527 UP000283509 UP000075883 UP000076858 UP000092461 UP000054359 UP000030765 UP000198287 UP000002358 UP000002320 UP000030742 UP000069272 UP000215335 UP000075901 UP000007266 UP000078540 UP000075884 UP000235965 UP000078542 UP000183832 UP000008237
UP000008792 UP000007798 UP000092445 UP000037069 UP000092553 UP000192221 UP000009192 UP000000673 UP000007801 UP000002282 UP000001070 UP000078200 UP000092443 UP000092460 UP000268350 UP000091820 UP000008711 UP000001819 UP000092444 UP000008744 UP000075885 UP000075903 UP000095301 UP000095300 UP000075882 UP000076407 UP000007062 UP000075840 UP000075880 UP000075881 UP000000803 UP000001292 UP000075886 UP000069940 UP000249989 UP000075902 UP000000305 UP000094527 UP000283509 UP000075883 UP000076858 UP000092461 UP000054359 UP000030765 UP000198287 UP000002358 UP000002320 UP000030742 UP000069272 UP000215335 UP000075901 UP000007266 UP000078540 UP000075884 UP000235965 UP000078542 UP000183832 UP000008237
Interpro
Gene 3D
ProteinModelPortal
H9JMA0
A0A2H1VZ67
A0A2A4K2S0
A0A194PHC7
A0A212EJD1
A0A0N0PEP3
+ More
I4DQ32 A0A0K8TT15 Q16W93 B4LTU9 A0A1B6LJ98 A0A2M3ZBW7 B4MW66 A0A2M4C0T0 A0A2M4C0V2 A0A023EGR2 A0A1A9ZIU1 A0A0L0C6Y4 A0A0M4DZT5 A0A1Q3FB65 A0A2S2NU69 A0A2M4AXR5 A0A2M4AXT8 A0A1W4VE14 B4KGC6 A0A2M4AXU6 A0A2M4AY26 A0A2S2QIK0 W5JDT0 B3MUA2 A0A1B6F3D4 B4P3K6 B4JAR7 A0A1A9VXL5 A0A1A9XDS5 A0A1B0B4M6 A0A034VH23 A0A3B0KB71 A0A1A9W1R4 C4WU99 B3N8W8 A0A1L8ECZ6 Q29MS9 D3TRG7 B4G8S6 A0A1L8E130 A0A1L8E106 A0A182PQC8 T1E8Y6 A0A2H8TM75 A0A182VCY0 A0A1I8MEE8 A0A1I8QCC3 A0A0K8VT04 A0A182KRE7 A0A182X3N7 A7URB7 A0A182I3D1 A0A182INU7 A0A182K442 Q9VQ35 B4IDB1 W8B2K8 A0A1Y1MN04 A0A2R7VX26 A0A0J9QTY8 A0A182QKH5 A0A182HBE7 A0A182U116 E9GLK0 A0A336MCA6 A0A1D2N342 A0A3R7PZX4 A0A182M046 A0A162CC62 A0A1B0GKG9 A0A0N8EJK3 A0A087T145 A0A0P5HU45 A0A084WND1 A0A226DAZ2 K7IUM6 B0WNR2 U4UPU0 A0A182FF66 A0A232FCT4 A0A182T7F3 A0A0P5HIN0 A0A0P5Q436 D2A319 A0A195BVM9 A0A182NHT2 A0A2J7RCX3 A0A195CPS4 A0A023G7C7 G3MS65 A0A023G4N8 A0A023FGC5 A0A1J1I9P5 E2B3D1
I4DQ32 A0A0K8TT15 Q16W93 B4LTU9 A0A1B6LJ98 A0A2M3ZBW7 B4MW66 A0A2M4C0T0 A0A2M4C0V2 A0A023EGR2 A0A1A9ZIU1 A0A0L0C6Y4 A0A0M4DZT5 A0A1Q3FB65 A0A2S2NU69 A0A2M4AXR5 A0A2M4AXT8 A0A1W4VE14 B4KGC6 A0A2M4AXU6 A0A2M4AY26 A0A2S2QIK0 W5JDT0 B3MUA2 A0A1B6F3D4 B4P3K6 B4JAR7 A0A1A9VXL5 A0A1A9XDS5 A0A1B0B4M6 A0A034VH23 A0A3B0KB71 A0A1A9W1R4 C4WU99 B3N8W8 A0A1L8ECZ6 Q29MS9 D3TRG7 B4G8S6 A0A1L8E130 A0A1L8E106 A0A182PQC8 T1E8Y6 A0A2H8TM75 A0A182VCY0 A0A1I8MEE8 A0A1I8QCC3 A0A0K8VT04 A0A182KRE7 A0A182X3N7 A7URB7 A0A182I3D1 A0A182INU7 A0A182K442 Q9VQ35 B4IDB1 W8B2K8 A0A1Y1MN04 A0A2R7VX26 A0A0J9QTY8 A0A182QKH5 A0A182HBE7 A0A182U116 E9GLK0 A0A336MCA6 A0A1D2N342 A0A3R7PZX4 A0A182M046 A0A162CC62 A0A1B0GKG9 A0A0N8EJK3 A0A087T145 A0A0P5HU45 A0A084WND1 A0A226DAZ2 K7IUM6 B0WNR2 U4UPU0 A0A182FF66 A0A232FCT4 A0A182T7F3 A0A0P5HIN0 A0A0P5Q436 D2A319 A0A195BVM9 A0A182NHT2 A0A2J7RCX3 A0A195CPS4 A0A023G7C7 G3MS65 A0A023G4N8 A0A023FGC5 A0A1J1I9P5 E2B3D1
PDB
6NU3
E-value=2.25678e-09,
Score=145
Ontologies
PANTHER
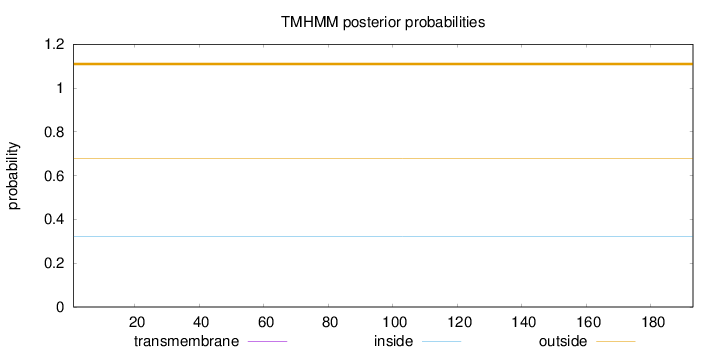
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00323
Exp number, first 60 AAs:
0.00182
Total prob of N-in:
0.32054
outside
1 - 193
Population Genetic Test Statistics
Pi
28.760568
Theta
24.866032
Tajima's D
0.493341
CLR
0
CSRT
0.52022398880056
Interpretation
Uncertain
