Pre Gene Modal
BGIBMGA010635
Annotation
PREDICTED:_mitochondrial_import_receptor_subunit_TOM70_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.585
Sequence
CDS
ATGGCTTCTACCGGTAGTACACCATTTCCGAAATGGCAATTGGCAATCCTTCTCGGTGCACCTATAGCTATCGGCTTAGGTTACCTTTACCTAAGAAATAGATTAGAAGACCCAGAAAAGAAGAAAGCATCAGAACTTAAAGCAAAAACGACAATATCTTTGGATAATGAAGAAAACACTACTAAGGTATCCGAAAGTGCCATAGAACGCGCCATGAAACTAAAAGGTGCTGGTAACCGGGCTTTTCATGCTGGAGAATATGACAAAGCCATTGCTTTATACAATGAAGCAATTGAAGCATGCCCCCCAGACCGTCCTGTCGATCTTGCTACATTTTACCAAAACCGCTCTGCTTGTTATGAAAAACGAGAAATGTGGGAACAAGTTAAAGAAGATTGCACTTTTGCATTAAAACTTAACGAAAAGTATGTAAAAGCATTTCTGCGTCGATCTCGTGCTGCGGAAAAAAGTGGCGATCTTGTGCTTGCTCTTGAAGATGTAACATCTGCATGTATCCTCGAGAGATTCCAAGTTCAGAGTTCCTTAGTAAATGCTGATCGTATTCTTAAAGCATTAGGCCGACAGCATGCCCGAGAAGCACTTGCTAAACGCAGCCCAGTCATGCCTTCAAAACATTTTATCAAAACATACTTTTCTGCATTCTCAGAGGACCCTATTGCAAAAATAAAAGTGGATGATGAAACTGATGGATTTGCTAAAGCTAGGAAAGCCCTTGACAATTTAGATTATGAATCAATTGTTGATGCTTGTACTGAAGAAATAGATGCAGAAGGAATTTTTAAAAATGAAGCTTTATTGCTGCGTGCAACCTTTTACCTATTGCTAGGTAGACATGAAGAAGCTCAGGCAGATCTTGGAAAAGTTATTGAAAGTGATGCTCCTACAAAAGTCAAAGTCAATGCTTTCATTAAACGAGCTTCACTTTTTACTCAGCTCGAAAATACAGAACGTTGCTTAGAAGATTTTGCTCAGGCCGCACAGCTAGATCCTAACAATTCTGACATTTATCATCACCGAGGACAAGTGTATTTATTATTAGAGAGAATGGATGAGGCAACTGCTGAATTTGCTAAAGCTGTAGAACTTAATCCAGATTTTTCCATTGCCTATATTCAAAAATGTTATGCTGACTATAGACATGCACAGATTAATAAGAATGTTGGTGCTTTGGTTCAAGTCAGAGCAGATTTTGAAAGTGCTTTGGAAAGATTTCCTAGATGTGCTGAAGCTTATATTTTGTATGCACAAGTTTTGTCAGATCAACAAGAGTGGGGTCGAGCTGAAGCATTGTTTGATGCTGCACTTGCTGTAGATCCAAATAATGCTACTCTGTATGTCCATAAGGGACTTGTTCAATTACAAAAGAGCACAGATTTTGATAAAGCTGTTAAATTGATAAATAAAGCAATCGAGATGGATGACAAATGTGACTTTGCCTATGAAACCTTGGGCACCATTGAAGTACAAAGGGGAAACCTAAGAAGATCACTAGAATTGTTTGAAAAGGCCATTGCATTAGCTAAAACTGAACTAGAAATGACTCATTTGTTTTCATTGAAAGATGCAGCTGCAGCTCAGCTCAAGGTCTCCGAGCGTTGGGGTCTAGACTGGACAGTGAGCTAA
Protein
MASTGSTPFPKWQLAILLGAPIAIGLGYLYLRNRLEDPEKKKASELKAKTTISLDNEENTTKVSESAIERAMKLKGAGNRAFHAGEYDKAIALYNEAIEACPPDRPVDLATFYQNRSACYEKREMWEQVKEDCTFALKLNEKYVKAFLRRSRAAEKSGDLVLALEDVTSACILERFQVQSSLVNADRILKALGRQHAREALAKRSPVMPSKHFIKTYFSAFSEDPIAKIKVDDETDGFAKARKALDNLDYESIVDACTEEIDAEGIFKNEALLLRATFYLLLGRHEEAQADLGKVIESDAPTKVKVNAFIKRASLFTQLENTERCLEDFAQAAQLDPNNSDIYHHRGQVYLLLERMDEATAEFAKAVELNPDFSIAYIQKCYADYRHAQINKNVGALVQVRADFESALERFPRCAEAYILYAQVLSDQQEWGRAEALFDAALAVDPNNATLYVHKGLVQLQKSTDFDKAVKLINKAIEMDDKCDFAYETLGTIEVQRGNLRRSLELFEKAIALAKTELEMTHLFSLKDAAAAQLKVSERWGLDWTVS
Summary
Uniprot
H9JM82
A0A2H1VZ49
A0A3S2TCA3
A0A2A4K348
A0A0L7KWY4
A0A194PI34
+ More
A0A0N1PKE8 A0A212EJC3 A0A067RFB3 A0A232FEY5 K7IZS8 U5EUC8 A0A1Y1K1D7 B0W5R7 A0A1Q3FI26 A0A1Q3FI34 Q0IF61 A0A0M9A839 A0A0T6AZT0 A0A158NBI1 A0A182GSA2 A0A023EV70 F4X8P1 A0A1J1HIM9 A0A195DSA8 A0A2A3EKM3 A0A195FC80 D2A1W3 A0A088A423 A0A162RV77 E0VRR7 A0A1L8DVS1 A0A0P5A6V0 A0A0P5Q9Z5 A0A0P6HUG8 A0A0P4WPR9 A0A151IGY8 A0A182PA76 H2TWD4 I3JK64 A0A3P8NE30 A0A3Q2WSH2 A0A3Q4HZP2 E2B022 T1IZN4 A0A2U9C938 A0A182JYJ7 A0A3S2N4Z5 A0A0P6BL06 F7BLU0 A0A2R9AC49 A0A2I3RQ35 A0A1B0DDC5 A0A2K5RKB4 A0A084VPI3 A0A3Q2E7Z0 A0A340X1T7 A0A383YMP2 A0A1A8AVU0 A0A1A8K9E6 A0A3B3D3X4 A0A3P9JEF9 A0A2I2ZI71 A0A3B4YRI4 H2MN90 A0A2K6T753 A0A1A8S2X8 A0A1A8M6V2 A0A1A8BDX9 A0A3Q1CFY7 A0A3P9MIK1 A0A146NSG7 A0A1A8GWD6 A0A2I3H3M1 A0A3B4TBC1 A0A3Q3B0R4 A0A1W4ZNX1 A0A182IE57 A0A1A7YKX4 A0A2U4ABD4 A0A2Y9FRB7 A0A3Q1EBJ6 A0A341CK44 A0A2Y9PNE3 U3AT82 G1SF32 A0A182TGB6 D2H5W0 A0A146X8W5 A0A2I4C6R8 A0A3B3WX24 A0A087YD40 A0A1V4J390 A0A3B5APB4 A0A0P7WX17 A0A182X0E6 A0A3P8TNP3 A0A2K5CA19 A0A2K5RKB8 A0A182RGD8
A0A0N1PKE8 A0A212EJC3 A0A067RFB3 A0A232FEY5 K7IZS8 U5EUC8 A0A1Y1K1D7 B0W5R7 A0A1Q3FI26 A0A1Q3FI34 Q0IF61 A0A0M9A839 A0A0T6AZT0 A0A158NBI1 A0A182GSA2 A0A023EV70 F4X8P1 A0A1J1HIM9 A0A195DSA8 A0A2A3EKM3 A0A195FC80 D2A1W3 A0A088A423 A0A162RV77 E0VRR7 A0A1L8DVS1 A0A0P5A6V0 A0A0P5Q9Z5 A0A0P6HUG8 A0A0P4WPR9 A0A151IGY8 A0A182PA76 H2TWD4 I3JK64 A0A3P8NE30 A0A3Q2WSH2 A0A3Q4HZP2 E2B022 T1IZN4 A0A2U9C938 A0A182JYJ7 A0A3S2N4Z5 A0A0P6BL06 F7BLU0 A0A2R9AC49 A0A2I3RQ35 A0A1B0DDC5 A0A2K5RKB4 A0A084VPI3 A0A3Q2E7Z0 A0A340X1T7 A0A383YMP2 A0A1A8AVU0 A0A1A8K9E6 A0A3B3D3X4 A0A3P9JEF9 A0A2I2ZI71 A0A3B4YRI4 H2MN90 A0A2K6T753 A0A1A8S2X8 A0A1A8M6V2 A0A1A8BDX9 A0A3Q1CFY7 A0A3P9MIK1 A0A146NSG7 A0A1A8GWD6 A0A2I3H3M1 A0A3B4TBC1 A0A3Q3B0R4 A0A1W4ZNX1 A0A182IE57 A0A1A7YKX4 A0A2U4ABD4 A0A2Y9FRB7 A0A3Q1EBJ6 A0A341CK44 A0A2Y9PNE3 U3AT82 G1SF32 A0A182TGB6 D2H5W0 A0A146X8W5 A0A2I4C6R8 A0A3B3WX24 A0A087YD40 A0A1V4J390 A0A3B5APB4 A0A0P7WX17 A0A182X0E6 A0A3P8TNP3 A0A2K5CA19 A0A2K5RKB8 A0A182RGD8
Pubmed
EMBL
BABH01033361
ODYU01005347
SOQ46098.1
RSAL01000420
RVE41802.1
NWSH01000241
+ More
PCG78072.1 JTDY01004948 KOB67564.1 KQ459603 KPI92713.1 KQ459752 KPJ20195.1 AGBW02014491 OWR41586.1 KK852723 KDR17690.1 NNAY01000319 OXU29262.1 GANO01002381 JAB57490.1 GEZM01095919 JAV55264.1 DS231844 EDS35720.1 GFDL01007851 JAV27194.1 GFDL01007828 JAV27217.1 CH477394 EAT41876.1 EAT41877.1 KQ435728 KOX77789.1 LJIG01022522 KRT80135.1 ADTU01011112 ADTU01011113 JXUM01084237 JXUM01084238 KQ563425 KXJ73854.1 GAPW01000696 JAC12902.1 GL888932 EGI57310.1 CVRI01000001 CRK86326.1 KQ980581 KYN15404.1 KZ288219 PBC32258.1 KQ981673 KYN38240.1 KQ971338 EFA02076.1 LRGB01000115 KZS20808.1 DS235494 EEB16073.1 GFDF01003543 JAV10541.1 GDIP01204175 JAJ19227.1 GDIQ01117249 JAL34477.1 GDIQ01014059 JAN80678.1 GDRN01072894 JAI63474.1 KQ977661 KYN00698.1 AERX01028549 AERX01055791 AERX01055792 GL444349 EFN60967.1 JH431720 CP026254 AWP11392.1 CM012440 RVE74085.1 GDIP01016489 JAM87226.1 AJFE02075596 AJFE02075597 AACZ04038560 AJVK01031827 AJVK01031828 ATLV01015007 KE524999 KFB39877.1 HADY01020664 SBP59149.1 HAED01016590 HAEE01008908 SBR28958.1 CABD030022985 HAEH01021604 HAEI01011152 SBS12596.1 HAEF01011278 HAEG01001368 SBR51989.1 HADZ01001831 HAEA01010634 SBP65772.1 GCES01152206 JAQ34116.1 HAEB01011520 HAEC01007231 SBQ75369.1 ADFV01005709 ADFV01005710 ADFV01005711 ADFV01005712 APCN01004569 HADW01017225 HADX01009035 SBP31267.1 GAMT01008479 GAMT01008478 GAMS01000942 GAMR01003200 GAMQ01001939 GAMQ01001938 GAMQ01001937 GAMQ01001936 GAMP01010550 JAB03383.1 JAB22194.1 JAB30732.1 JAB39912.1 JAB42205.1 AAGW02016179 AAGW02016180 AAGW02016181 AAGW02016182 GL192515 EFB30312.1 GCES01047720 JAR38603.1 AYCK01012527 LSYS01009367 OPJ66691.1 JARO02004351 KPP68670.1
PCG78072.1 JTDY01004948 KOB67564.1 KQ459603 KPI92713.1 KQ459752 KPJ20195.1 AGBW02014491 OWR41586.1 KK852723 KDR17690.1 NNAY01000319 OXU29262.1 GANO01002381 JAB57490.1 GEZM01095919 JAV55264.1 DS231844 EDS35720.1 GFDL01007851 JAV27194.1 GFDL01007828 JAV27217.1 CH477394 EAT41876.1 EAT41877.1 KQ435728 KOX77789.1 LJIG01022522 KRT80135.1 ADTU01011112 ADTU01011113 JXUM01084237 JXUM01084238 KQ563425 KXJ73854.1 GAPW01000696 JAC12902.1 GL888932 EGI57310.1 CVRI01000001 CRK86326.1 KQ980581 KYN15404.1 KZ288219 PBC32258.1 KQ981673 KYN38240.1 KQ971338 EFA02076.1 LRGB01000115 KZS20808.1 DS235494 EEB16073.1 GFDF01003543 JAV10541.1 GDIP01204175 JAJ19227.1 GDIQ01117249 JAL34477.1 GDIQ01014059 JAN80678.1 GDRN01072894 JAI63474.1 KQ977661 KYN00698.1 AERX01028549 AERX01055791 AERX01055792 GL444349 EFN60967.1 JH431720 CP026254 AWP11392.1 CM012440 RVE74085.1 GDIP01016489 JAM87226.1 AJFE02075596 AJFE02075597 AACZ04038560 AJVK01031827 AJVK01031828 ATLV01015007 KE524999 KFB39877.1 HADY01020664 SBP59149.1 HAED01016590 HAEE01008908 SBR28958.1 CABD030022985 HAEH01021604 HAEI01011152 SBS12596.1 HAEF01011278 HAEG01001368 SBR51989.1 HADZ01001831 HAEA01010634 SBP65772.1 GCES01152206 JAQ34116.1 HAEB01011520 HAEC01007231 SBQ75369.1 ADFV01005709 ADFV01005710 ADFV01005711 ADFV01005712 APCN01004569 HADW01017225 HADX01009035 SBP31267.1 GAMT01008479 GAMT01008478 GAMS01000942 GAMR01003200 GAMQ01001939 GAMQ01001938 GAMQ01001937 GAMQ01001936 GAMP01010550 JAB03383.1 JAB22194.1 JAB30732.1 JAB39912.1 JAB42205.1 AAGW02016179 AAGW02016180 AAGW02016181 AAGW02016182 GL192515 EFB30312.1 GCES01047720 JAR38603.1 AYCK01012527 LSYS01009367 OPJ66691.1 JARO02004351 KPP68670.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000037510
UP000053268
UP000053240
+ More
UP000007151 UP000027135 UP000215335 UP000002358 UP000002320 UP000008820 UP000053105 UP000005205 UP000069940 UP000249989 UP000007755 UP000183832 UP000078492 UP000242457 UP000078541 UP000007266 UP000005203 UP000076858 UP000009046 UP000078542 UP000075885 UP000005226 UP000005207 UP000265100 UP000264840 UP000261580 UP000000311 UP000246464 UP000075881 UP000008225 UP000240080 UP000002277 UP000092462 UP000233040 UP000030765 UP000265020 UP000265300 UP000261681 UP000261560 UP000265200 UP000001519 UP000261360 UP000001038 UP000233220 UP000257160 UP000265180 UP000001073 UP000261420 UP000264800 UP000192224 UP000075840 UP000245320 UP000248484 UP000257200 UP000252040 UP000248483 UP000001811 UP000075902 UP000265000 UP000192220 UP000261480 UP000028760 UP000190648 UP000261400 UP000034805 UP000076407 UP000265080 UP000233020 UP000075900
UP000007151 UP000027135 UP000215335 UP000002358 UP000002320 UP000008820 UP000053105 UP000005205 UP000069940 UP000249989 UP000007755 UP000183832 UP000078492 UP000242457 UP000078541 UP000007266 UP000005203 UP000076858 UP000009046 UP000078542 UP000075885 UP000005226 UP000005207 UP000265100 UP000264840 UP000261580 UP000000311 UP000246464 UP000075881 UP000008225 UP000240080 UP000002277 UP000092462 UP000233040 UP000030765 UP000265020 UP000265300 UP000261681 UP000261560 UP000265200 UP000001519 UP000261360 UP000001038 UP000233220 UP000257160 UP000265180 UP000001073 UP000261420 UP000264800 UP000192224 UP000075840 UP000245320 UP000248484 UP000257200 UP000252040 UP000248483 UP000001811 UP000075902 UP000265000 UP000192220 UP000261480 UP000028760 UP000190648 UP000261400 UP000034805 UP000076407 UP000265080 UP000233020 UP000075900
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JM82
A0A2H1VZ49
A0A3S2TCA3
A0A2A4K348
A0A0L7KWY4
A0A194PI34
+ More
A0A0N1PKE8 A0A212EJC3 A0A067RFB3 A0A232FEY5 K7IZS8 U5EUC8 A0A1Y1K1D7 B0W5R7 A0A1Q3FI26 A0A1Q3FI34 Q0IF61 A0A0M9A839 A0A0T6AZT0 A0A158NBI1 A0A182GSA2 A0A023EV70 F4X8P1 A0A1J1HIM9 A0A195DSA8 A0A2A3EKM3 A0A195FC80 D2A1W3 A0A088A423 A0A162RV77 E0VRR7 A0A1L8DVS1 A0A0P5A6V0 A0A0P5Q9Z5 A0A0P6HUG8 A0A0P4WPR9 A0A151IGY8 A0A182PA76 H2TWD4 I3JK64 A0A3P8NE30 A0A3Q2WSH2 A0A3Q4HZP2 E2B022 T1IZN4 A0A2U9C938 A0A182JYJ7 A0A3S2N4Z5 A0A0P6BL06 F7BLU0 A0A2R9AC49 A0A2I3RQ35 A0A1B0DDC5 A0A2K5RKB4 A0A084VPI3 A0A3Q2E7Z0 A0A340X1T7 A0A383YMP2 A0A1A8AVU0 A0A1A8K9E6 A0A3B3D3X4 A0A3P9JEF9 A0A2I2ZI71 A0A3B4YRI4 H2MN90 A0A2K6T753 A0A1A8S2X8 A0A1A8M6V2 A0A1A8BDX9 A0A3Q1CFY7 A0A3P9MIK1 A0A146NSG7 A0A1A8GWD6 A0A2I3H3M1 A0A3B4TBC1 A0A3Q3B0R4 A0A1W4ZNX1 A0A182IE57 A0A1A7YKX4 A0A2U4ABD4 A0A2Y9FRB7 A0A3Q1EBJ6 A0A341CK44 A0A2Y9PNE3 U3AT82 G1SF32 A0A182TGB6 D2H5W0 A0A146X8W5 A0A2I4C6R8 A0A3B3WX24 A0A087YD40 A0A1V4J390 A0A3B5APB4 A0A0P7WX17 A0A182X0E6 A0A3P8TNP3 A0A2K5CA19 A0A2K5RKB8 A0A182RGD8
A0A0N1PKE8 A0A212EJC3 A0A067RFB3 A0A232FEY5 K7IZS8 U5EUC8 A0A1Y1K1D7 B0W5R7 A0A1Q3FI26 A0A1Q3FI34 Q0IF61 A0A0M9A839 A0A0T6AZT0 A0A158NBI1 A0A182GSA2 A0A023EV70 F4X8P1 A0A1J1HIM9 A0A195DSA8 A0A2A3EKM3 A0A195FC80 D2A1W3 A0A088A423 A0A162RV77 E0VRR7 A0A1L8DVS1 A0A0P5A6V0 A0A0P5Q9Z5 A0A0P6HUG8 A0A0P4WPR9 A0A151IGY8 A0A182PA76 H2TWD4 I3JK64 A0A3P8NE30 A0A3Q2WSH2 A0A3Q4HZP2 E2B022 T1IZN4 A0A2U9C938 A0A182JYJ7 A0A3S2N4Z5 A0A0P6BL06 F7BLU0 A0A2R9AC49 A0A2I3RQ35 A0A1B0DDC5 A0A2K5RKB4 A0A084VPI3 A0A3Q2E7Z0 A0A340X1T7 A0A383YMP2 A0A1A8AVU0 A0A1A8K9E6 A0A3B3D3X4 A0A3P9JEF9 A0A2I2ZI71 A0A3B4YRI4 H2MN90 A0A2K6T753 A0A1A8S2X8 A0A1A8M6V2 A0A1A8BDX9 A0A3Q1CFY7 A0A3P9MIK1 A0A146NSG7 A0A1A8GWD6 A0A2I3H3M1 A0A3B4TBC1 A0A3Q3B0R4 A0A1W4ZNX1 A0A182IE57 A0A1A7YKX4 A0A2U4ABD4 A0A2Y9FRB7 A0A3Q1EBJ6 A0A341CK44 A0A2Y9PNE3 U3AT82 G1SF32 A0A182TGB6 D2H5W0 A0A146X8W5 A0A2I4C6R8 A0A3B3WX24 A0A087YD40 A0A1V4J390 A0A3B5APB4 A0A0P7WX17 A0A182X0E6 A0A3P8TNP3 A0A2K5CA19 A0A2K5RKB8 A0A182RGD8
PDB
3FP4
E-value=1.00927e-27,
Score=308
Ontologies
GO
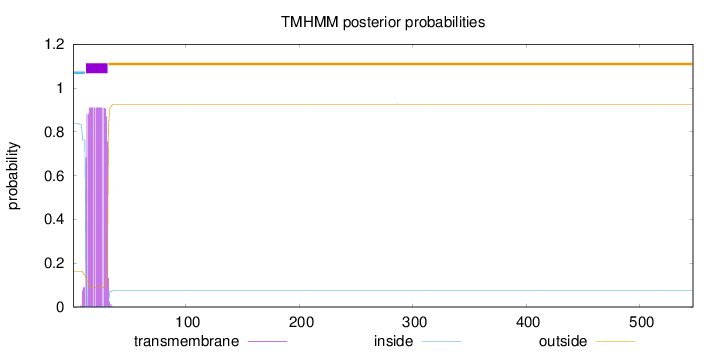
Topology
Length:
547
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.17449
Exp number, first 60 AAs:
18.17158
Total prob of N-in:
0.83741
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 31
outside
32 - 547
Population Genetic Test Statistics
Pi
13.611375
Theta
15.351584
Tajima's D
0.494389
CLR
0
CSRT
0.517874106294685
Interpretation
Uncertain
