Gene
KWMTBOMO07291
Pre Gene Modal
BGIBMGA010651
Annotation
PREDICTED:_coiled-coil_domain-containing_protein_97_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.846
Sequence
CDS
ATGGAAAAAATCAACATTAATTTAAAAAATAAATTCAACGAAAGTGAAGTGAATCCTGTAACTGATATCATTGACTACCTAGTAAGAAGTGCAAATATATCCTTTAAGAATTCCCATGCTAATGAACAACCGGTAAGGACATGTGAGAAACTGCAAATGGCCCTTGAAATTTATAAAAGATCACCTGTAGAATTCCTCATGCAATTCGGAAAATATTTAGCTCCTAATCACATAAAATACTTTGAAAATATCTCTAGCTCCAAAGATCAAACATTTAGGAATTACGTGCACTATTTGAGTGACTATCATTCGGAAACAAGCAGACGCAAGAGGATAAAGAACAGAAGATATAAGGCATTACAAAAAATGAAGATTAAAACTGATTATTTTAGTGAAAAACAAATGATGTACAGAAATCCTCTTTTATATGAACAGTTGGTTGGGCAATATTTAACAGATGAAGAAATAAAAGAAAGAGATCGAATAACTGAAAACGCCAGTTTTCTTAACTTAATATTGGAAACTGTAGATCACAACCACATGAGAGAGGTGAAGAATGTGCAAATGCTCGAAGAAGGTATTCATGAAACACCAGAAGCTTCTGAATTTTCTTTGAATGATAGTACTAATGAAAACATAAATTTTCAACACAAAAGAAAATGGGGTGAATTCACTACCCCAGATACGAAACCTGACTATATTCCAGAAAACAGAAAACACAGAGTTATTAGTGCTCCAGAAAAGAAGTTATTGTATGATGAGTTCCTTCAAGAAATGTATAGCAGCTTTATAGATGGTAATGATATTGACATTGATTATAAAACTATTGATAATGATGAGCAGTATGATGATTTAGAGCAGGAGTCACAAGATGCAGAAGATAAATATTTTGATTCTGAAACAAATGATGTTGAGAACTTAGAAGAACACATGCACCTTGTAGAGGAATATGGTCAAAGTAAAATAAATAATCTTGAAAAAGATGACCCTCTTGATGTGTTTATGGAGCATATATCTAAGAAGTTTCAACTGAATTAA
Protein
MEKININLKNKFNESEVNPVTDIIDYLVRSANISFKNSHANEQPVRTCEKLQMALEIYKRSPVEFLMQFGKYLAPNHIKYFENISSSKDQTFRNYVHYLSDYHSETSRRKRIKNRRYKALQKMKIKTDYFSEKQMMYRNPLLYEQLVGQYLTDEEIKERDRITENASFLNLILETVDHNHMREVKNVQMLEEGIHETPEASEFSLNDSTNENINFQHKRKWGEFTTPDTKPDYIPENRKHRVISAPEKKLLYDEFLQEMYSSFIDGNDIDIDYKTIDNDEQYDDLEQESQDAEDKYFDSETNDVENLEEHMHLVEEYGQSKINNLEKDDPLDVFMEHISKKFQLN
Summary
Uniprot
H9JM98
A0A2H1VZ38
A0A2A4K2D0
A0A1E1W6Z7
A0A0L7KZN6
A0A0N1ICV7
+ More
A0A194PH38 D1ZZF4 A0A1A9VMG3 A0A195E671 A0A1B0BL94 A0A151XKH2 A0A0L7QJK2 A0A1A9Y3K3 N6UV90 J3JYG9 A0A158NLJ3 A0A195B1H4 A0A1A9WM48 A0A195EY81 A0A026WZ82 F4WW32 A0A3L8DRA9 E9IEN1 A0A0J7KYX2 A0A151IB51 A0A1B6ECE4 A0A154PDG1 A0A336KTA6 A0A0N1ITG9 A0A0L0CFR8 A0A2A3EEH7 A0A0A1WKD2 A0A1I8Q2D1 A0A182MHZ3 A0A1B6EWJ6 A0A1I8MQE2 A0A2P8ZL26 A0A1W4XJG1 A0A084WGW0 A0A182RZU4 Q176Q2 A0A182GG10 A0A067R222 E2B0Q3 A0A069DSB3 A0A1B6LXK8 A0A182W743 A0A0K8UW39 A0A182JGF8 B4JBW2 A0A0M4EF13 A0A224XGT6 E2BGH6 A0A1Q3FDD1 A0A2J7RC46 A0A0V0GCU5 A0A0K8WEV5 B4LSH7 B4KJM9 A0A1W4V8T2 W5J7N4 W8CBQ5 A0A182NUC7 B4G901 A0A0P4VRH9 A0A182QBS3 A0A1Y1K0S1 Q9VLP6 B0X6R1 B4HYI3 A0A1S4K432 B4NWV9 V5IEQ7 A0A2R5LIH5 A0A182V271 B4MTJ4 T1PNJ3 A0A087ZYP0 B3N738 A0A182JQX0 B4Q6Q1 A0A034WBQ0 Q7Q3Z2 B3MPG4 B7Q0F4 A0A1S4GY03 A0A182L1I3 A0A310SS24 A0A182TWR8 A0A182F4U5 G3MR78 A0A0L8IA04 A0A182WU38 A0A182HUQ0 A0A0K8STY9 A0A1B0CZD9 H3BF82
A0A194PH38 D1ZZF4 A0A1A9VMG3 A0A195E671 A0A1B0BL94 A0A151XKH2 A0A0L7QJK2 A0A1A9Y3K3 N6UV90 J3JYG9 A0A158NLJ3 A0A195B1H4 A0A1A9WM48 A0A195EY81 A0A026WZ82 F4WW32 A0A3L8DRA9 E9IEN1 A0A0J7KYX2 A0A151IB51 A0A1B6ECE4 A0A154PDG1 A0A336KTA6 A0A0N1ITG9 A0A0L0CFR8 A0A2A3EEH7 A0A0A1WKD2 A0A1I8Q2D1 A0A182MHZ3 A0A1B6EWJ6 A0A1I8MQE2 A0A2P8ZL26 A0A1W4XJG1 A0A084WGW0 A0A182RZU4 Q176Q2 A0A182GG10 A0A067R222 E2B0Q3 A0A069DSB3 A0A1B6LXK8 A0A182W743 A0A0K8UW39 A0A182JGF8 B4JBW2 A0A0M4EF13 A0A224XGT6 E2BGH6 A0A1Q3FDD1 A0A2J7RC46 A0A0V0GCU5 A0A0K8WEV5 B4LSH7 B4KJM9 A0A1W4V8T2 W5J7N4 W8CBQ5 A0A182NUC7 B4G901 A0A0P4VRH9 A0A182QBS3 A0A1Y1K0S1 Q9VLP6 B0X6R1 B4HYI3 A0A1S4K432 B4NWV9 V5IEQ7 A0A2R5LIH5 A0A182V271 B4MTJ4 T1PNJ3 A0A087ZYP0 B3N738 A0A182JQX0 B4Q6Q1 A0A034WBQ0 Q7Q3Z2 B3MPG4 B7Q0F4 A0A1S4GY03 A0A182L1I3 A0A310SS24 A0A182TWR8 A0A182F4U5 G3MR78 A0A0L8IA04 A0A182WU38 A0A182HUQ0 A0A0K8STY9 A0A1B0CZD9 H3BF82
Pubmed
19121390
26227816
26354079
18362917
19820115
23537049
+ More
22516182 21347285 24508170 21719571 30249741 21282665 26108605 25830018 25315136 29403074 24438588 17510324 26483478 24845553 20798317 26334808 17994087 20920257 23761445 24495485 27129103 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25765539 22936249 25348373 12364791 20966253 22216098 9215903
22516182 21347285 24508170 21719571 30249741 21282665 26108605 25830018 25315136 29403074 24438588 17510324 26483478 24845553 20798317 26334808 17994087 20920257 23761445 24495485 27129103 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25765539 22936249 25348373 12364791 20966253 22216098 9215903
EMBL
BABH01033361
ODYU01005347
SOQ46101.1
NWSH01000241
PCG78088.1
GDQN01008433
+ More
JAT82621.1 JTDY01004181 KOB68494.1 KQ459752 KPJ20192.1 KQ459603 KPI92716.1 KQ971338 EFA01860.1 KQ979579 KYN20668.1 JXJN01016272 KQ982038 KYQ60788.1 LHQN01050120 KOC58793.1 APGK01009864 APGK01024888 APGK01029864 KB740749 KB740565 KB738057 ENN79071.1 ENN80223.1 ENN82797.1 BT128297 KB632221 AEE63255.1 ERL90195.1 ADTU01019550 ADTU01019551 KQ976662 KYM78316.1 KQ981914 KYN33106.1 KK107063 EZA61103.1 GL888404 EGI61621.1 QOIP01000005 RLU22419.1 GL762653 EFZ20971.1 LBMM01001796 KMQ95737.1 KQ978125 KYM96884.1 GEDC01001689 JAS35609.1 KQ434878 KZC09883.1 UFQS01000899 UFQT01000899 SSX07567.1 SSX27907.1 KQ435804 KOX73032.1 JRES01000536 KNC30334.1 KZ288269 PBC29874.1 GBXI01015297 JAC98994.1 AXCM01002772 GECZ01027394 JAS42375.1 PYGN01000025 PSN57208.1 ATLV01023763 KE525346 KFB49454.1 CH477384 EAT42140.1 JXUM01060755 KQ562117 KXJ76649.1 KK852758 KDR17052.1 GL444666 EFN60725.1 GBGD01002342 JAC86547.1 GEBQ01011594 GEBQ01009130 JAT28383.1 JAT30847.1 GDHF01021422 JAI30892.1 CH916368 EDW04065.1 CP012523 ALC38905.1 GFTR01004720 JAW11706.1 GL448189 EFN85169.1 GFDL01009502 JAV25543.1 NEVH01005887 PNF38402.1 GECL01000403 JAP05721.1 GDHF01002682 JAI49632.1 CH940649 EDW64799.1 CH933807 EDW11474.1 ADMH02002111 ETN58885.1 GAMC01000274 JAC06282.1 CH479180 EDW28831.1 GDKW01002949 JAI53646.1 AXCN02000765 GEZM01100369 GEZM01100368 GEZM01100367 GEZM01100366 JAV52916.1 AE014134 AY119623 AAF52638.1 AAM50277.1 DS232421 EDS41544.1 CH480818 EDW52113.1 CM000157 EDW88489.1 GANP01009767 JAB74701.1 GGLE01005198 MBY09324.1 CH963852 EDW75433.2 KA650284 AFP64913.1 CH954177 EDV59334.1 CM000361 CM002910 EDX04210.1 KMY89036.1 GAKP01005936 JAC53016.1 AAAB01008964 EAA12696.3 CH902620 EDV31260.1 ABJB010527341 ABJB010576251 ABJB010848096 ABJB011061716 DS831942 EEC12326.1 KQ761010 OAD58394.1 JO844379 AEO35996.1 KQ416252 KOF97850.1 APCN01002471 GBRD01009058 GBRD01000714 JAG56763.1 AJVK01009542 AFYH01010020
JAT82621.1 JTDY01004181 KOB68494.1 KQ459752 KPJ20192.1 KQ459603 KPI92716.1 KQ971338 EFA01860.1 KQ979579 KYN20668.1 JXJN01016272 KQ982038 KYQ60788.1 LHQN01050120 KOC58793.1 APGK01009864 APGK01024888 APGK01029864 KB740749 KB740565 KB738057 ENN79071.1 ENN80223.1 ENN82797.1 BT128297 KB632221 AEE63255.1 ERL90195.1 ADTU01019550 ADTU01019551 KQ976662 KYM78316.1 KQ981914 KYN33106.1 KK107063 EZA61103.1 GL888404 EGI61621.1 QOIP01000005 RLU22419.1 GL762653 EFZ20971.1 LBMM01001796 KMQ95737.1 KQ978125 KYM96884.1 GEDC01001689 JAS35609.1 KQ434878 KZC09883.1 UFQS01000899 UFQT01000899 SSX07567.1 SSX27907.1 KQ435804 KOX73032.1 JRES01000536 KNC30334.1 KZ288269 PBC29874.1 GBXI01015297 JAC98994.1 AXCM01002772 GECZ01027394 JAS42375.1 PYGN01000025 PSN57208.1 ATLV01023763 KE525346 KFB49454.1 CH477384 EAT42140.1 JXUM01060755 KQ562117 KXJ76649.1 KK852758 KDR17052.1 GL444666 EFN60725.1 GBGD01002342 JAC86547.1 GEBQ01011594 GEBQ01009130 JAT28383.1 JAT30847.1 GDHF01021422 JAI30892.1 CH916368 EDW04065.1 CP012523 ALC38905.1 GFTR01004720 JAW11706.1 GL448189 EFN85169.1 GFDL01009502 JAV25543.1 NEVH01005887 PNF38402.1 GECL01000403 JAP05721.1 GDHF01002682 JAI49632.1 CH940649 EDW64799.1 CH933807 EDW11474.1 ADMH02002111 ETN58885.1 GAMC01000274 JAC06282.1 CH479180 EDW28831.1 GDKW01002949 JAI53646.1 AXCN02000765 GEZM01100369 GEZM01100368 GEZM01100367 GEZM01100366 JAV52916.1 AE014134 AY119623 AAF52638.1 AAM50277.1 DS232421 EDS41544.1 CH480818 EDW52113.1 CM000157 EDW88489.1 GANP01009767 JAB74701.1 GGLE01005198 MBY09324.1 CH963852 EDW75433.2 KA650284 AFP64913.1 CH954177 EDV59334.1 CM000361 CM002910 EDX04210.1 KMY89036.1 GAKP01005936 JAC53016.1 AAAB01008964 EAA12696.3 CH902620 EDV31260.1 ABJB010527341 ABJB010576251 ABJB010848096 ABJB011061716 DS831942 EEC12326.1 KQ761010 OAD58394.1 JO844379 AEO35996.1 KQ416252 KOF97850.1 APCN01002471 GBRD01009058 GBRD01000714 JAG56763.1 AJVK01009542 AFYH01010020
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007266
+ More
UP000078200 UP000078492 UP000092460 UP000075809 UP000053825 UP000092443 UP000019118 UP000030742 UP000005205 UP000078540 UP000091820 UP000078541 UP000053097 UP000007755 UP000279307 UP000036403 UP000078542 UP000076502 UP000053105 UP000037069 UP000242457 UP000095300 UP000075883 UP000095301 UP000245037 UP000192223 UP000030765 UP000075900 UP000008820 UP000069940 UP000249989 UP000027135 UP000000311 UP000075920 UP000075880 UP000001070 UP000092553 UP000008237 UP000235965 UP000008792 UP000009192 UP000192221 UP000000673 UP000075884 UP000008744 UP000075886 UP000000803 UP000002320 UP000001292 UP000002282 UP000075903 UP000007798 UP000005203 UP000008711 UP000075881 UP000000304 UP000007062 UP000007801 UP000001555 UP000075882 UP000075902 UP000069272 UP000053454 UP000076407 UP000075840 UP000092462 UP000008672
UP000078200 UP000078492 UP000092460 UP000075809 UP000053825 UP000092443 UP000019118 UP000030742 UP000005205 UP000078540 UP000091820 UP000078541 UP000053097 UP000007755 UP000279307 UP000036403 UP000078542 UP000076502 UP000053105 UP000037069 UP000242457 UP000095300 UP000075883 UP000095301 UP000245037 UP000192223 UP000030765 UP000075900 UP000008820 UP000069940 UP000249989 UP000027135 UP000000311 UP000075920 UP000075880 UP000001070 UP000092553 UP000008237 UP000235965 UP000008792 UP000009192 UP000192221 UP000000673 UP000075884 UP000008744 UP000075886 UP000000803 UP000002320 UP000001292 UP000002282 UP000075903 UP000007798 UP000005203 UP000008711 UP000075881 UP000000304 UP000007062 UP000007801 UP000001555 UP000075882 UP000075902 UP000069272 UP000053454 UP000076407 UP000075840 UP000092462 UP000008672
ProteinModelPortal
H9JM98
A0A2H1VZ38
A0A2A4K2D0
A0A1E1W6Z7
A0A0L7KZN6
A0A0N1ICV7
+ More
A0A194PH38 D1ZZF4 A0A1A9VMG3 A0A195E671 A0A1B0BL94 A0A151XKH2 A0A0L7QJK2 A0A1A9Y3K3 N6UV90 J3JYG9 A0A158NLJ3 A0A195B1H4 A0A1A9WM48 A0A195EY81 A0A026WZ82 F4WW32 A0A3L8DRA9 E9IEN1 A0A0J7KYX2 A0A151IB51 A0A1B6ECE4 A0A154PDG1 A0A336KTA6 A0A0N1ITG9 A0A0L0CFR8 A0A2A3EEH7 A0A0A1WKD2 A0A1I8Q2D1 A0A182MHZ3 A0A1B6EWJ6 A0A1I8MQE2 A0A2P8ZL26 A0A1W4XJG1 A0A084WGW0 A0A182RZU4 Q176Q2 A0A182GG10 A0A067R222 E2B0Q3 A0A069DSB3 A0A1B6LXK8 A0A182W743 A0A0K8UW39 A0A182JGF8 B4JBW2 A0A0M4EF13 A0A224XGT6 E2BGH6 A0A1Q3FDD1 A0A2J7RC46 A0A0V0GCU5 A0A0K8WEV5 B4LSH7 B4KJM9 A0A1W4V8T2 W5J7N4 W8CBQ5 A0A182NUC7 B4G901 A0A0P4VRH9 A0A182QBS3 A0A1Y1K0S1 Q9VLP6 B0X6R1 B4HYI3 A0A1S4K432 B4NWV9 V5IEQ7 A0A2R5LIH5 A0A182V271 B4MTJ4 T1PNJ3 A0A087ZYP0 B3N738 A0A182JQX0 B4Q6Q1 A0A034WBQ0 Q7Q3Z2 B3MPG4 B7Q0F4 A0A1S4GY03 A0A182L1I3 A0A310SS24 A0A182TWR8 A0A182F4U5 G3MR78 A0A0L8IA04 A0A182WU38 A0A182HUQ0 A0A0K8STY9 A0A1B0CZD9 H3BF82
A0A194PH38 D1ZZF4 A0A1A9VMG3 A0A195E671 A0A1B0BL94 A0A151XKH2 A0A0L7QJK2 A0A1A9Y3K3 N6UV90 J3JYG9 A0A158NLJ3 A0A195B1H4 A0A1A9WM48 A0A195EY81 A0A026WZ82 F4WW32 A0A3L8DRA9 E9IEN1 A0A0J7KYX2 A0A151IB51 A0A1B6ECE4 A0A154PDG1 A0A336KTA6 A0A0N1ITG9 A0A0L0CFR8 A0A2A3EEH7 A0A0A1WKD2 A0A1I8Q2D1 A0A182MHZ3 A0A1B6EWJ6 A0A1I8MQE2 A0A2P8ZL26 A0A1W4XJG1 A0A084WGW0 A0A182RZU4 Q176Q2 A0A182GG10 A0A067R222 E2B0Q3 A0A069DSB3 A0A1B6LXK8 A0A182W743 A0A0K8UW39 A0A182JGF8 B4JBW2 A0A0M4EF13 A0A224XGT6 E2BGH6 A0A1Q3FDD1 A0A2J7RC46 A0A0V0GCU5 A0A0K8WEV5 B4LSH7 B4KJM9 A0A1W4V8T2 W5J7N4 W8CBQ5 A0A182NUC7 B4G901 A0A0P4VRH9 A0A182QBS3 A0A1Y1K0S1 Q9VLP6 B0X6R1 B4HYI3 A0A1S4K432 B4NWV9 V5IEQ7 A0A2R5LIH5 A0A182V271 B4MTJ4 T1PNJ3 A0A087ZYP0 B3N738 A0A182JQX0 B4Q6Q1 A0A034WBQ0 Q7Q3Z2 B3MPG4 B7Q0F4 A0A1S4GY03 A0A182L1I3 A0A310SS24 A0A182TWR8 A0A182F4U5 G3MR78 A0A0L8IA04 A0A182WU38 A0A182HUQ0 A0A0K8STY9 A0A1B0CZD9 H3BF82
Ontologies
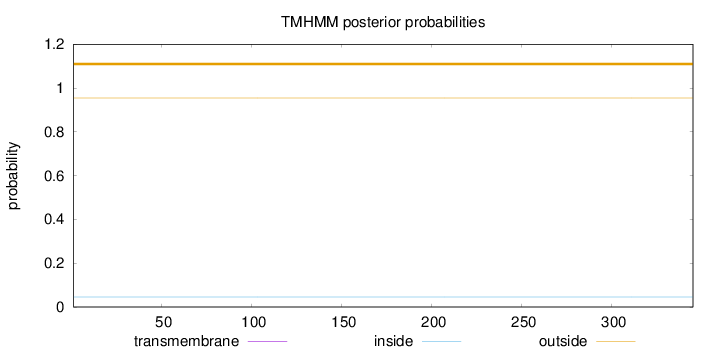
Topology
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04572
outside
1 - 345
Population Genetic Test Statistics
Pi
20.479647
Theta
17.677773
Tajima's D
0.470771
CLR
0.394965
CSRT
0.51762411879406
Interpretation
Uncertain
