Gene
KWMTBOMO07282
Pre Gene Modal
BGIBMGA010639
Annotation
PREDICTED:_lipase_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.42 PlasmaMembrane Reliability : 1.077
Sequence
CDS
ATGAAGTGGTTGTTAGGATTGTTTTTCTGTATATGTGTTAATCAGGCGACCGCTCGGCGTTCTCCACATGCGGACTACGTCGAGGAACTGTACAGGTTGGACGCCCAAGGTGCCCGTTATTCTACCAATGTAACTGAAGATGCTTTGCTAGATATTGTTGGACTTATCAAAAAATATAGATATCCAGTAGAAGTTCACAAGGTCGTTACGGAAGATGGCTATATCTTAGAAATGCAAAGAATTCCTCATGGACGCGACCAGAACAACCGTCCTGACCCGAAGAAACCAGCTGTTTTAGTTATGCATGGACTGTTTGCTTCTGCTGCTGACTGGGTTTTGATGGGTCCAGGTGTTGGTTTAGCATACATTTTGGCTGAAGCTGGTTATGATGTGTGGCTTGGAAATGCACGTGGTACATATTACTCCAGAGCTCACATTAAATTAGACCCGGACAACGACTCAGAGTTTTGGAAATTCAGTTGGGAAGAAATAGGTACCCGAGATCTGCCTGCTATGATTGATTACACTTTGAAAGTTGCTGGAAAAAGACGACTTCATTATATTGGTCACTCACAAGGAACTACTGTCTTTTGGGCTATGGGATCTTTGAGACCTGAATACAACAGCAAAATTATTGCTATGCAAGCTTATGCTCCCGTAGCTTACTTAGAATTTAATGCAAACAGATTACTTAAACTCATTGCTCCACATGCCAACAGTATTGAGGCACTTACAAGTCTTATTGGAATAAATGAGCTATTCGGAAGATCTGACTTCTTTACCAACTTGGGAATGAGATTCTGCGCTGATGGCACATTTTTCCAGGCGATGTGCACAAATATGTTATTCGCTTTTGCCGGACGCAGTGAAGACATGCACAATGCCACTATGTTGCCTGTGAAACTAGGACATACACCTGCAGGCATTGCTGTTAGACAGATAGCACATTACGGACAACTTATCAACAAAAATTTCTTCAGACGATATGATCACGGACGCTTGACAAATTGGAGGGTTTACAGAAGTTTCACACCGCCAAGATATGACCTGTCTCTCATAACCGCCCCTGTTTTTCTACACTATGTTGACGAGGACATCTTTGCTGATGATGTCTATGGCTTCAGCGGTATCCTGAGGAGACGGGGTAGAGTCCGGGCTAAACCGGAGCGATGGTGGATTAGATTCAGCCAGACTGATGCCCAGCGTGTGCCAATCCACCACAGTCCTCGTGCCGGGAACGGAATCGGGAGCGAGACCGAGCTTCATAACGACGGGACAGTGGCCCCGGAGAGTGCCACCTGTCGATGCCCATATCATGGTTGGAGGCGTCCGTATCGAGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCATTCTGGACAGTCGTTGGACCTTCCGTGCTCACTTTCAGAACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTGCCCAACTTGGCAGAAGCCGGCTTAACGCGCCTGACCTGGGGGGCCTGACCAGCCCCAGTGTGGTCAGAGCGCGGAGGGCCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGACGGCTGGCCGATCCTTCAGCTGGTCGTAGGACCGTCGAGGCGATTTGCCCGGTTCTTGTGAATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGCTTCGGTGAGTTCCTGCACCGGATCGGAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTCGCGACTTGGACACGGCGGAGCACACGCACACACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGATCCGACGGTTGGGGCTCGAGGTGGCGCTCGACAAAACCCAGGCCCTGTTATTTCACGGGCCGGGACGAGCGCCGCCTCTGGGTGCCCACCTCGTGATCGGAGGTGTCCGCGTCGGGGTCGGGGTGACCGGTCTTCGGTACCTCGGCCTCGAACTGGACGGTCGGTGGAGCTTCCGCGCTCACTTCGAAAAATTGGGCCCTCGGCTGATGGCGACGGCCGGCTCTCTGAGCCGGCTTCTTCCAAACGTCGGGGGTCCCGATCAGGTGGCGCGCCGCCTCTACATGGGGGTGGTGCGGTCGATGGCACTATACGGCGCTCCCGTATGGTGCCACGCCCTGACCCGCGAGAACGTCGCGGCGCTGCGACGTCCGCAGCGCGCGATCGCGGTCAGGGCGATCCGAGGATACCGCACCGTCTCCTTCGAGGCGGCGTGCTTGCTCGCCGGGGCGCCACCCTGGGACCTGGAGGCGGAGGCGCTCGCTGCCGATTACCGGTGGCGTAGCGACCTTCGCTCTAGGGGGGAAGGGCGCCCCGGCGAAGGTGTAGTTCGAGCGCGGAGGCTCCACTCTCGGCGGTCCGTGCTGGAGGCGTGGTCTCGCCGCTTGGCGGACCCGTCGGCCGGCCTCCGAACCGTCGAGGCGGTCCGCCCGGTTCTCGCGGACTGGGTGGGCCGCGACCGCGGATGCCTCACCTTCCGGCTAACGCAGATGCTTACCGGCCATGGTTGTTTCGGCCGGTACTTGCACAAGATAGCCGGAAGGGAACCGACGGCGCAGTGCCACCACTGCGCGGACCGCGACGAGGAAGACACAGCGGAACACACGCTGGCGCGTTGCTCTGGATTCGACGAGCAACGCGCCGCCCTCGTCGCGGTCATTGGAGAGGACCTCTCGCTGCCGCGCGTCGTGGCTACGATGCTCGGCAGCGACGCGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTCCACCATATCGCAGAAGGAGGCGGCGGAGCGAGAGAGGGAGAGCTCTTCCCTTTCCGCACCGATCCGTCGCCGTCGAGCCGGGGGCCGGAGGCGGGAATACGCCCGTACGTCCCGGCCCCTGTAG
Protein
MKWLLGLFFCICVNQATARRSPHADYVEELYRLDAQGARYSTNVTEDALLDIVGLIKKYRYPVEVHKVVTEDGYILEMQRIPHGRDQNNRPDPKKPAVLVMHGLFASAADWVLMGPGVGLAYILAEAGYDVWLGNARGTYYSRAHIKLDPDNDSEFWKFSWEEIGTRDLPAMIDYTLKVAGKRRLHYIGHSQGTTVFWAMGSLRPEYNSKIIAMQAYAPVAYLEFNANRLLKLIAPHANSIEALTSLIGINELFGRSDFFTNLGMRFCADGTFFQAMCTNMLFAFAGRSEDMHNATMLPVKLGHTPAGIAVRQIAHYGQLINKNFFRRYDHGRLTNWRVYRSFTPPRYDLSLITAPVFLHYVDEDIFADDVYGFSGILRRRGRVRAKPERWWIRFSQTDAQRVPIHHSPRAGNGIGSETELHNDGTVAPESATCRCPYHGWRRPYRGRGAVEVPRPHSGQSLDLPCSLSEPGPSFVGGGRRVKPASAQLGRSRLNAPDLGGLTSPSVVRARRAQSRRSVLESWSRRLADPSAGRRTVEAICPVLVNWVNRDRGRLTFRLTQVLTGHGCFGEFLHRIGAEPTAECHHCGRDLDTAEHTHTLVVAECCGIDARIRRLGLEVALDKTQALLFHGPGRAPPLGAHLVIGGVRVGVGVTGLRYLGLELDGRWSFRAHFEKLGPRLMATAGSLSRLLPNVGGPDQVARRLYMGVVRSMALYGAPVWCHALTRENVAALRRPQRAIAVRAIRGYRTVSFEAACLLAGAPPWDLEAEALAADYRWRSDLRSRGEGRPGEGVVRARRLHSRRSVLEAWSRRLADPSAGLRTVEAVRPVLADWVGRDRGCLTFRLTQMLTGHGCFGRYLHKIAGREPTAQCHHCADRDEEDTAEHTLARCSGFDEQRAALVAVIGEDLSLPRVVATMLGSDASWKAMLDFCESTISQKEAAERERESSSLSAPIRRRRAGGRRREYARTSRPL
Summary
Uniprot
ProteinModelPortal
PDB
1HLG
E-value=2.95634e-57,
Score=566
Ontologies
PATHWAY
GO
Topology
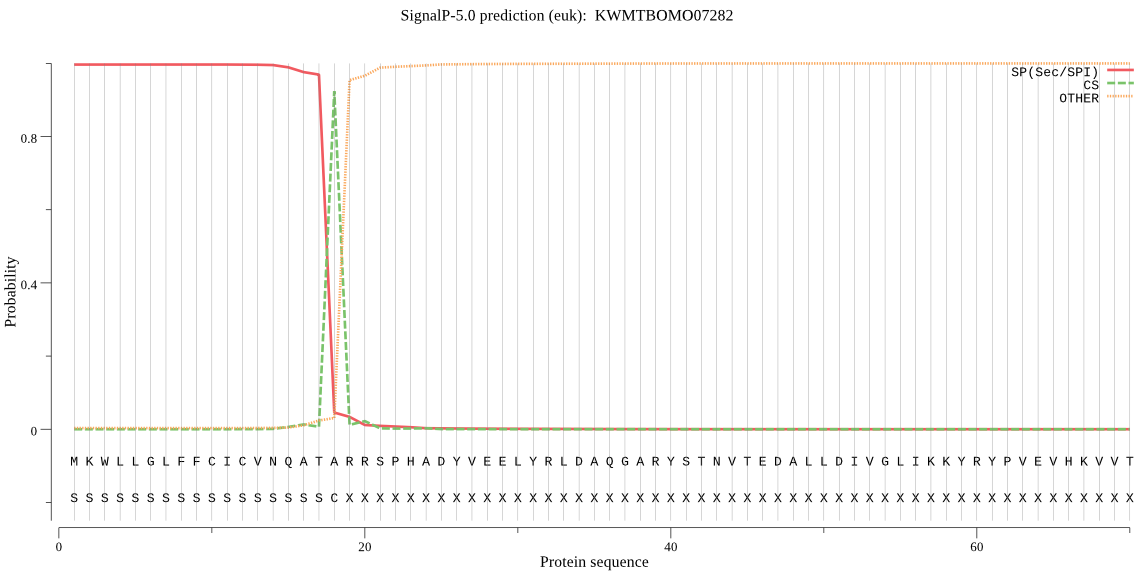
SignalP
Position: 1 - 18,
Likelihood: 0.996387
Length:
973
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.59664
Exp number, first 60 AAs:
0.00236
Total prob of N-in:
0.01350
outside
1 - 973

Population Genetic Test Statistics
Pi
0.256674
Theta
1.243726
Tajima's D
-1.427822
CLR
2.3117
CSRT
0.0577971101444928
Interpretation
Uncertain
