Gene
KWMTBOMO07274 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010644
Annotation
aubergine_protein_[Bombyx_mori]
Full name
Piwi-like protein Siwi
Location in the cell
Nuclear Reliability : 2.152
Sequence
CDS
ATGTCAGAACCGAGAGGTAGAGGACGAGCTCGAGGACGCGCTGGTCGTGGTGGTGATGGAGGCGGACCAGCACCACGACGTCCAGGGGAACAAGCAGGCCCATCACAACAATCTATGCCACCTGGTCCTAGACCTCAACCTCCATCTGGCTGGGGACCACAATCCAGTGTTCCTCCAGTAAGAGCTGGTGTCCCGACACCAACAGCACAAGCGGGTCGCGCCTCACACCGTGTTACTCCCACTACACATGAACATCCAGGAGATATCGATGTGCAACAACGCATGCAAAAATTAGAACTGGGACCCCATTCATCTGGAGGTGGTGATGCATCATCCGTAGTTGGCCGTGGCTCTCGACGAGGGGGTGGAAGAGTCCTTCCTGAAACTATTTCCATCTTGCGCACACGTCCAGAAGCTGTAACATCTAAGAAAGGTACTTCGGGTACTCCCCTGGATCTGCTTGCCAACTATTTTACTGTTGAAACAACTCCCAAATGGGGCCTGTATCAATATCACGTCGATATTTCTCCAGAAGAAGACAGCACTGGAGTCCGAAAGGCACTGATGCGTGTTCACAGCAAAACACTCGGAGGATATCTCTTTGATGGCACTGTGTTGTACACTGTAAATCGTCTGCATCCTGATCCGATGGAGCTATATTCTGATCGTAAAACGGATAACGAACGTATGCGAATCTTAATTAAGCTGACCTGCGAAGTTAGCCCTGGAGACTATCACTATATACAAATCTTTAACATCATTATAAGGAAATGCTTTAATTTATTGAAGCTACAGCTTATGGGACGGGATTATTTTGACCCTGAAGCAAAGATTGATATACCAGAATTCAAGCTTCAAATTTGGCCAGGATATAAAACGACAATCAATCAATATGAAGATCGTTTGTTGTTGGTCACTGAAATCGCCCATAAGGTGCTTCGCATGGACACTGTTCTTCAAATGTTGAGTGAATATGCCGCAACAAAAGGCAATAATTACAAAAAAATATTTTTAGAAGATGTTGTTGGAAAAATTGTAATGACTGACTACAATAAAAGAACTTACAGAGTTGACGATGTAGCTTGGAATGTGTCGCCGAAGTCCACATTTAAAATGCGTGACGAAAACATTACCTATATTGAATACTATTATAAGAAATACAATCTACGCATTCAAGATCCGGGTCAACCGCTTCTGATTTCGCGGTCGAAGCCGCGTGAGATTCGTGCGGGTCTGCCTGAGTTGATATATCTAGTGCCAGAACTGTGCCGTCAGACCGGCCTCTCCGACGAGATGCGAGCTAATTTCAAGCTGATGCGCTCATTGGACGTCCATACAAAGATTGGGCCCGACAAGCGCATTGAAAAGCTTAATAATTTCAACAGGAGATTCACAAGCACACCTGAAGTTGTTGAAGAACTGGCAACATGGTCGTTGAAACTATCTAAGGAACTGGTAAAAATAAAGGGTCGCCAACTTCCACCTGAAAATATAATACAAGCAAATAATGTTAAATACCCTGCAGGAGACACCACCGAGGGCTGGACGCGAGATATGAGATCGAAGCATCTCCTGGCTATTGCCCAACTAAACTCTTGGGTAGTGATCACCCCAGAAAGACAACGCCGTGATACAGAGTCGTTCATTGATCTCATAATAAAAACTGGCGGAGGTGTTGGTTTTCGAATGAGAAGCCCTGACTTAGTTGTGATTCGTCATGACGGTCCGATAGAATACGCTAATATGTGTGAAGAAGTGATTGCAAGGAAAAATCCTGCCCTGATACTGTGCGTTCTGGCTAGGAATTATGCTGATAGGTATGAAGCCATAAAAAAGAAATGCACTGTAGACCGAGCCGTGCCGACGCAGGTAGTCTGCGCTCGTAACATGAGTAGCAAGTCGGCTATGTCAATTGCTACAAAAGTTGCTATTCAAATAAACTGTAAGCTTGGGGGTTCTCCGTGGACAGTAGATATCCCACTGCCGTCACTGATGGTGGTCGGGTACGACGTGTGCCACGACACTCGTTCGAAAGAAAAGAGCTTCGGGGCCTTCGTAGCCACCCTCGACAAACAGATGACTCAGTACTATTCAATCGTCAATGCGCACACTTCCGGAGAAGAACTCAGCTCTCACATGGGTTTCAATATCGCGTCCGCTGTCAAAAAATTCCGCGAGAAAAACGGGACCTACCCAGCCCGAATCTTCATTTACCGCGACGGTGTCGGTGATGGTCAAATCCCATACGTGCATAGCCATGAAGTTGCTGAGATAAAAAAGAAATTGGCAGAAATTTATGCAGGAGTCGAAATAAAGTTGGCATTCATTATTGTGTCGAAACGTATTAACACCCGAATCTTCGTTCAACGTGGTCGCAGCGGAGAAAATCCACGCCCTGGCACAGTGATTGACGACGTCGTCACCTTGCCTGAACGGTATGATTTCTATCTGGTTTCACAAAATGTACGTGAAGGTACCATTGCTCCTACCTCATACAATGTGATTGAAGATACAACAGGACTCAATCCTGATCGTATCCAACGTTTGACCTACAAACTGACACACTTGTATTTCAACTGCTCTTCCCAAGTTCGCGTCCCATCGGTCTGCCAATATGCCCACAAACTTGCATTCCTTGCAGCTAACAGTCTGCACAACCAACCGCATTATTCATTGAATGAAACTTTATATTTCCTCTAA
Protein
MSEPRGRGRARGRAGRGGDGGGPAPRRPGEQAGPSQQSMPPGPRPQPPSGWGPQSSVPPVRAGVPTPTAQAGRASHRVTPTTHEHPGDIDVQQRMQKLELGPHSSGGGDASSVVGRGSRRGGGRVLPETISILRTRPEAVTSKKGTSGTPLDLLANYFTVETTPKWGLYQYHVDISPEEDSTGVRKALMRVHSKTLGGYLFDGTVLYTVNRLHPDPMELYSDRKTDNERMRILIKLTCEVSPGDYHYIQIFNIIIRKCFNLLKLQLMGRDYFDPEAKIDIPEFKLQIWPGYKTTINQYEDRLLLVTEIAHKVLRMDTVLQMLSEYAATKGNNYKKIFLEDVVGKIVMTDYNKRTYRVDDVAWNVSPKSTFKMRDENITYIEYYYKKYNLRIQDPGQPLLISRSKPREIRAGLPELIYLVPELCRQTGLSDEMRANFKLMRSLDVHTKIGPDKRIEKLNNFNRRFTSTPEVVEELATWSLKLSKELVKIKGRQLPPENIIQANNVKYPAGDTTEGWTRDMRSKHLLAIAQLNSWVVITPERQRRDTESFIDLIIKTGGGVGFRMRSPDLVVIRHDGPIEYANMCEEVIARKNPALILCVLARNYADRYEAIKKKCTVDRAVPTQVVCARNMSSKSAMSIATKVAIQINCKLGGSPWTVDIPLPSLMVVGYDVCHDTRSKEKSFGAFVATLDKQMTQYYSIVNAHTSGEELSSHMGFNIASAVKKFREKNGTYPARIFIYRDGVGDGQIPYVHSHEVAEIKKKLAEIYAGVEIKLAFIIVSKRINTRIFVQRGRSGENPRPGTVIDDVVTLPERYDFYLVSQNVREGTIAPTSYNVIEDTTGLNPDRIQRLTYKLTHLYFNCSSQVRVPSVCQYAHKLAFLAANSLHNQPHYSLNETLYFL
Summary
Description
Endoribonuclease that plays a central role during spermatogenesis by repressing transposable elements and preventing their mobilization, which is essential for the germline integrity (PubMed:19460866, PubMed:27693359). Plays an essential role in meiotic differentiation of spermatocytes, germ cell differentiation and in self-renewal of spermatogonial stem cells (PubMed:19460866, PubMed:25558067, PubMed:27693359). Its presence in oocytes suggests that it may participate in similar functions during oogenesis in females (PubMed:18191035). Acts via the piRNA metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and govern the methylation and subsequent repression of transposons (PubMed:19460866, PubMed:25558067, PubMed:27693359). Directly binds piRNAs, a class of 24 to 30 nucleotide RNAs that are generated by a Dicer-independent mechanism and are primarily derived from transposons and other repeated sequence elements (PubMed:19460866, PubMed:25558067, PubMed:27693359). Recognizes piRNAs containing a phosphate at the 5'-end and a 2'-O-methylation modification at the 3'-end (PubMed:27693359). Strongly prefers a uridine in the first position of their guide (g1U preference, also named 1U-bias) and a complementary adenosine in the target (t1A bias) (PubMed:24757166, PubMed:27693359). Plays a key role in the piRNA amplification loop, also named ping-pong amplification cycle: antisense piRNA-bound Siwi and sense piRNA-bound Ago3 reciprocally cleave complementary transcripts, to couple the amplification of piRNAs with the repression of transposable elements. In this process Siwi acts as a 'slicer-competent' piRNA endoribonuclease that cleaves primary piRNAs, which are then loaded onto Ago3 (PubMed:27693359). In this process, Siwi requires the RNA unwinding activity of the RNA helicase Vasa for the release of the cleavage products (PubMed:25558067).
Cofactor
Mg(2+)
Subunit
Interacts (when symmetrically methylated) with Papi/TDRKH (PubMed:23970546, PubMed:26919431). Interacts with Vasa (PubMed:25558067).
Similarity
Belongs to the argonaute family. Piwi subfamily.
Belongs to the argonaute family.
Belongs to the argonaute family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Developmental protein
Differentiation
Endonuclease
Hydrolase
Meiosis
Metal-binding
Methylation
Nuclease
Reference proteome
RNA-binding
RNA-mediated gene silencing
Spermatogenesis
Feature
chain Piwi-like protein Siwi
Uniprot
A8D8P8
A0A3S2N9X4
A0A2A4J8B1
A0A2H1VJ96
S4PEK0
A0A194R5H6
+ More
A0A0K0K804 A0A194PIH2 A0A2H1V8Y3 A0A2H1V5K6 A0A212FEI4 U5IBZ2 A0A067RM87 S4W045 A0A1Y1N8W6 A0A1L7NZM7 A0A2J7RAL3 K4MR11 A0A0T6B2M2 A0A1B6KR98 A0A1B6MJY0 A0A1B6LZB3 A0A0L7RCR7 A0A1B6F8K3 K7ISV0 A0A1W4XLK3 V5I9K0 A0A232ES36 A0A1B6I4E4 A0A088ARC0 A0A2I6PIY2 A0A026WTT9 A0A1B6DVK7 D6WSL8 A0A0J7L598 A0A1B6DT98 A0A1B6CSA2 C9E0T3 C9E0T4 A0A232FMW2 E2AFM7 A0A151II10 A0A195ED42 A0A0P4VWF7 A0A067RW77 A0A151XBT8 K7IP24 A0A0C9RXH0 A0A224X701 A0A023F683 A0A0C9Q8U0 A0A195FXG8 A0A158NV03 A0A195B8E8 A0A069DWG2 A0A146L4W1 A0A0V0G407 A0A1B6CDN8 F4WNM9 E2ALX7 K7J555 A0A1B6DLF2 A0A1L7NZN1 A0A336MED6 A0A336MWF3 A0A232FJQ3 A0A195CQF2 A0A126G917 E0W0U4 A0A1B6GNL7 A0A195EMS4 A0A0N0U5A5 A0A158NCK2 A0A336L7R3 A0A195BXG1 A0A336MSM0 A0A195F956 A0A0A9Z7D5 A0A2S2R8W0 A0A2J7QB22 F4WX64 J9JYB0 Q16IF2 A0A182GB18 Q16ZS3 A0A1S4FZX7 E2BM63 M4QXX0 A0A1S4J618 B0W5T3 B0W5P0 J9JWH1 J9K744 A0A1I8PZG7 A0A1I8PZE8 A0A151X5U6 A0A1Q3FC13 A0A1Q3G0H1 A0A182G925 A0A1S4J6G4
A0A0K0K804 A0A194PIH2 A0A2H1V8Y3 A0A2H1V5K6 A0A212FEI4 U5IBZ2 A0A067RM87 S4W045 A0A1Y1N8W6 A0A1L7NZM7 A0A2J7RAL3 K4MR11 A0A0T6B2M2 A0A1B6KR98 A0A1B6MJY0 A0A1B6LZB3 A0A0L7RCR7 A0A1B6F8K3 K7ISV0 A0A1W4XLK3 V5I9K0 A0A232ES36 A0A1B6I4E4 A0A088ARC0 A0A2I6PIY2 A0A026WTT9 A0A1B6DVK7 D6WSL8 A0A0J7L598 A0A1B6DT98 A0A1B6CSA2 C9E0T3 C9E0T4 A0A232FMW2 E2AFM7 A0A151II10 A0A195ED42 A0A0P4VWF7 A0A067RW77 A0A151XBT8 K7IP24 A0A0C9RXH0 A0A224X701 A0A023F683 A0A0C9Q8U0 A0A195FXG8 A0A158NV03 A0A195B8E8 A0A069DWG2 A0A146L4W1 A0A0V0G407 A0A1B6CDN8 F4WNM9 E2ALX7 K7J555 A0A1B6DLF2 A0A1L7NZN1 A0A336MED6 A0A336MWF3 A0A232FJQ3 A0A195CQF2 A0A126G917 E0W0U4 A0A1B6GNL7 A0A195EMS4 A0A0N0U5A5 A0A158NCK2 A0A336L7R3 A0A195BXG1 A0A336MSM0 A0A195F956 A0A0A9Z7D5 A0A2S2R8W0 A0A2J7QB22 F4WX64 J9JYB0 Q16IF2 A0A182GB18 Q16ZS3 A0A1S4FZX7 E2BM63 M4QXX0 A0A1S4J618 B0W5T3 B0W5P0 J9JWH1 J9K744 A0A1I8PZG7 A0A1I8PZE8 A0A151X5U6 A0A1Q3FC13 A0A1Q3G0H1 A0A182G925 A0A1S4J6G4
EC Number
3.1.26.-
Pubmed
18191035
19121390
17698031
19460866
23970546
24757166
+ More
25558067 26919431 27693359 23622113 26354079 22118469 24845553 23792802 28004739 28034629 23122849 20075255 28648823 29267401 24508170 30249741 18362917 19820115 20513057 20798317 27129103 25474469 21347285 26334808 26823975 21719571 26868596 20566863 25401762 17510324 26483478
25558067 26919431 27693359 23622113 26354079 22118469 24845553 23792802 28004739 28034629 23122849 20075255 28648823 29267401 24508170 30249741 18362917 19820115 20513057 20798317 27129103 25474469 21347285 26334808 26823975 21719571 26868596 20566863 25401762 17510324 26483478
EMBL
AB332313
AB372006
EU143547
BABH01033344
EU034629
RSAL01000375
+ More
RVE42096.1 NWSH01002708 PCG67642.1 ODYU01002684 SOQ40502.1 GAIX01004467 JAA88093.1 KQ460883 KPJ11116.1 JX481277 AGS40930.1 KQ459603 KPI92848.1 ODYU01001293 SOQ37310.1 ODYU01000777 SOQ36079.1 AGBW02008943 OWR52118.1 JX998175 AFW04603.1 KK852427 KDR24113.1 KF032645 AGO85970.1 GEZM01010669 JAV93928.1 LC194148 BAW35370.1 NEVH01006567 PNF37866.1 JQ434103 AFV31611.1 LJIG01016089 KRT81648.1 GEBQ01026026 JAT13951.1 GEBQ01003782 JAT36195.1 GEBQ01010950 JAT29027.1 KQ414615 KOC68594.1 GECZ01023197 GECZ01007473 JAS46572.1 JAS62296.1 AAZX01003322 GALX01002800 JAB65666.1 NNAY01002506 OXU21173.1 GECU01025898 JAS81808.1 MG266017 AUM60044.1 KK107109 QOIP01000008 EZA59378.1 RLU19528.1 GEDC01007582 JAS29716.1 KQ971354 EFA07425.1 LBMM01000695 KMQ97723.1 GEDC01008396 JAS28902.1 GEDC01021000 JAS16298.1 GQ444142 ACV84377.1 GQ444143 ACV84378.1 NNAY01000022 OXU31859.1 GL439118 EFN67778.1 KQ977588 KYN01664.1 KQ979074 KYN22764.1 GDKW01001568 JAI55027.1 KDR24114.1 KQ982316 KYQ57823.1 GBYB01012591 JAG82358.1 GFTR01008201 JAW08225.1 GBBI01001987 JAC16725.1 GBYB01010683 JAG80450.1 KQ981215 KYN44539.1 ADTU01002871 ADTU01002872 KQ976565 KYM80517.1 GBGD01000526 JAC88363.1 GDHC01016829 JAQ01800.1 GECL01003336 JAP02788.1 GEDC01025741 JAS11557.1 GL888237 EGI64222.1 GL440682 EFN65569.1 AAZX01015855 GEDC01010784 JAS26514.1 LC194149 BAW35371.1 UFQT01001059 SSX28665.1 UFQT01003030 SSX34460.1 NNAY01000117 OXU30813.1 KQ977408 KYN02941.1 KR012681 ALC79956.1 DS235862 EEB19250.1 GECZ01031744 GECZ01005733 JAS38025.1 JAS64036.1 KQ978691 KYN29202.1 KQ435791 KOX74374.1 ADTU01011951 UFQS01002193 UFQT01002193 SSX13459.1 SSX32890.1 KQ976396 KYM92985.1 UFQT01002105 SSX32695.1 KQ981727 KYN36973.1 GBHO01004331 GBHO01004330 GBRD01000007 GDHC01012215 GDHC01005575 GDHC01000298 JAG39273.1 JAG39274.1 JAG65814.1 JAQ06414.1 JAQ13054.1 JAQ18331.1 GGMS01017205 MBY86408.1 NEVH01016302 PNF25798.1 GL888417 EGI61307.1 ABLF02030598 CH478081 EAT34046.1 JXUM01157421 JXUM01157422 KQ572743 KXJ68003.1 CH477485 EAT40148.1 GL449158 EFN83189.1 KC316041 AGH30330.1 DS231844 EDS35736.1 EDS35693.1 ABLF02019072 ABLF02019073 ABLF02066598 ABLF02038523 ABLF02038526 KQ982490 KYQ55785.1 GFDL01009929 JAV25116.1 GFDL01001779 JAV33266.1 JXUM01153240 KQ571658 KXJ68137.1
RVE42096.1 NWSH01002708 PCG67642.1 ODYU01002684 SOQ40502.1 GAIX01004467 JAA88093.1 KQ460883 KPJ11116.1 JX481277 AGS40930.1 KQ459603 KPI92848.1 ODYU01001293 SOQ37310.1 ODYU01000777 SOQ36079.1 AGBW02008943 OWR52118.1 JX998175 AFW04603.1 KK852427 KDR24113.1 KF032645 AGO85970.1 GEZM01010669 JAV93928.1 LC194148 BAW35370.1 NEVH01006567 PNF37866.1 JQ434103 AFV31611.1 LJIG01016089 KRT81648.1 GEBQ01026026 JAT13951.1 GEBQ01003782 JAT36195.1 GEBQ01010950 JAT29027.1 KQ414615 KOC68594.1 GECZ01023197 GECZ01007473 JAS46572.1 JAS62296.1 AAZX01003322 GALX01002800 JAB65666.1 NNAY01002506 OXU21173.1 GECU01025898 JAS81808.1 MG266017 AUM60044.1 KK107109 QOIP01000008 EZA59378.1 RLU19528.1 GEDC01007582 JAS29716.1 KQ971354 EFA07425.1 LBMM01000695 KMQ97723.1 GEDC01008396 JAS28902.1 GEDC01021000 JAS16298.1 GQ444142 ACV84377.1 GQ444143 ACV84378.1 NNAY01000022 OXU31859.1 GL439118 EFN67778.1 KQ977588 KYN01664.1 KQ979074 KYN22764.1 GDKW01001568 JAI55027.1 KDR24114.1 KQ982316 KYQ57823.1 GBYB01012591 JAG82358.1 GFTR01008201 JAW08225.1 GBBI01001987 JAC16725.1 GBYB01010683 JAG80450.1 KQ981215 KYN44539.1 ADTU01002871 ADTU01002872 KQ976565 KYM80517.1 GBGD01000526 JAC88363.1 GDHC01016829 JAQ01800.1 GECL01003336 JAP02788.1 GEDC01025741 JAS11557.1 GL888237 EGI64222.1 GL440682 EFN65569.1 AAZX01015855 GEDC01010784 JAS26514.1 LC194149 BAW35371.1 UFQT01001059 SSX28665.1 UFQT01003030 SSX34460.1 NNAY01000117 OXU30813.1 KQ977408 KYN02941.1 KR012681 ALC79956.1 DS235862 EEB19250.1 GECZ01031744 GECZ01005733 JAS38025.1 JAS64036.1 KQ978691 KYN29202.1 KQ435791 KOX74374.1 ADTU01011951 UFQS01002193 UFQT01002193 SSX13459.1 SSX32890.1 KQ976396 KYM92985.1 UFQT01002105 SSX32695.1 KQ981727 KYN36973.1 GBHO01004331 GBHO01004330 GBRD01000007 GDHC01012215 GDHC01005575 GDHC01000298 JAG39273.1 JAG39274.1 JAG65814.1 JAQ06414.1 JAQ13054.1 JAQ18331.1 GGMS01017205 MBY86408.1 NEVH01016302 PNF25798.1 GL888417 EGI61307.1 ABLF02030598 CH478081 EAT34046.1 JXUM01157421 JXUM01157422 KQ572743 KXJ68003.1 CH477485 EAT40148.1 GL449158 EFN83189.1 KC316041 AGH30330.1 DS231844 EDS35736.1 EDS35693.1 ABLF02019072 ABLF02019073 ABLF02066598 ABLF02038523 ABLF02038526 KQ982490 KYQ55785.1 GFDL01009929 JAV25116.1 GFDL01001779 JAV33266.1 JXUM01153240 KQ571658 KXJ68137.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000027135 UP000235965 UP000053825 UP000002358 UP000192223 UP000215335 UP000005203 UP000053097 UP000279307 UP000007266 UP000036403 UP000000311 UP000078542 UP000078492 UP000075809 UP000078541 UP000005205 UP000078540 UP000007755 UP000009046 UP000053105 UP000007819 UP000008820 UP000069940 UP000249989 UP000008237 UP000002320 UP000095300
UP000027135 UP000235965 UP000053825 UP000002358 UP000192223 UP000215335 UP000005203 UP000053097 UP000279307 UP000007266 UP000036403 UP000000311 UP000078542 UP000078492 UP000075809 UP000078541 UP000005205 UP000078540 UP000007755 UP000009046 UP000053105 UP000007819 UP000008820 UP000069940 UP000249989 UP000008237 UP000002320 UP000095300
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A8D8P8
A0A3S2N9X4
A0A2A4J8B1
A0A2H1VJ96
S4PEK0
A0A194R5H6
+ More
A0A0K0K804 A0A194PIH2 A0A2H1V8Y3 A0A2H1V5K6 A0A212FEI4 U5IBZ2 A0A067RM87 S4W045 A0A1Y1N8W6 A0A1L7NZM7 A0A2J7RAL3 K4MR11 A0A0T6B2M2 A0A1B6KR98 A0A1B6MJY0 A0A1B6LZB3 A0A0L7RCR7 A0A1B6F8K3 K7ISV0 A0A1W4XLK3 V5I9K0 A0A232ES36 A0A1B6I4E4 A0A088ARC0 A0A2I6PIY2 A0A026WTT9 A0A1B6DVK7 D6WSL8 A0A0J7L598 A0A1B6DT98 A0A1B6CSA2 C9E0T3 C9E0T4 A0A232FMW2 E2AFM7 A0A151II10 A0A195ED42 A0A0P4VWF7 A0A067RW77 A0A151XBT8 K7IP24 A0A0C9RXH0 A0A224X701 A0A023F683 A0A0C9Q8U0 A0A195FXG8 A0A158NV03 A0A195B8E8 A0A069DWG2 A0A146L4W1 A0A0V0G407 A0A1B6CDN8 F4WNM9 E2ALX7 K7J555 A0A1B6DLF2 A0A1L7NZN1 A0A336MED6 A0A336MWF3 A0A232FJQ3 A0A195CQF2 A0A126G917 E0W0U4 A0A1B6GNL7 A0A195EMS4 A0A0N0U5A5 A0A158NCK2 A0A336L7R3 A0A195BXG1 A0A336MSM0 A0A195F956 A0A0A9Z7D5 A0A2S2R8W0 A0A2J7QB22 F4WX64 J9JYB0 Q16IF2 A0A182GB18 Q16ZS3 A0A1S4FZX7 E2BM63 M4QXX0 A0A1S4J618 B0W5T3 B0W5P0 J9JWH1 J9K744 A0A1I8PZG7 A0A1I8PZE8 A0A151X5U6 A0A1Q3FC13 A0A1Q3G0H1 A0A182G925 A0A1S4J6G4
A0A0K0K804 A0A194PIH2 A0A2H1V8Y3 A0A2H1V5K6 A0A212FEI4 U5IBZ2 A0A067RM87 S4W045 A0A1Y1N8W6 A0A1L7NZM7 A0A2J7RAL3 K4MR11 A0A0T6B2M2 A0A1B6KR98 A0A1B6MJY0 A0A1B6LZB3 A0A0L7RCR7 A0A1B6F8K3 K7ISV0 A0A1W4XLK3 V5I9K0 A0A232ES36 A0A1B6I4E4 A0A088ARC0 A0A2I6PIY2 A0A026WTT9 A0A1B6DVK7 D6WSL8 A0A0J7L598 A0A1B6DT98 A0A1B6CSA2 C9E0T3 C9E0T4 A0A232FMW2 E2AFM7 A0A151II10 A0A195ED42 A0A0P4VWF7 A0A067RW77 A0A151XBT8 K7IP24 A0A0C9RXH0 A0A224X701 A0A023F683 A0A0C9Q8U0 A0A195FXG8 A0A158NV03 A0A195B8E8 A0A069DWG2 A0A146L4W1 A0A0V0G407 A0A1B6CDN8 F4WNM9 E2ALX7 K7J555 A0A1B6DLF2 A0A1L7NZN1 A0A336MED6 A0A336MWF3 A0A232FJQ3 A0A195CQF2 A0A126G917 E0W0U4 A0A1B6GNL7 A0A195EMS4 A0A0N0U5A5 A0A158NCK2 A0A336L7R3 A0A195BXG1 A0A336MSM0 A0A195F956 A0A0A9Z7D5 A0A2S2R8W0 A0A2J7QB22 F4WX64 J9JYB0 Q16IF2 A0A182GB18 Q16ZS3 A0A1S4FZX7 E2BM63 M4QXX0 A0A1S4J618 B0W5T3 B0W5P0 J9JWH1 J9K744 A0A1I8PZG7 A0A1I8PZE8 A0A151X5U6 A0A1Q3FC13 A0A1Q3G0H1 A0A182G925 A0A1S4J6G4
PDB
5GUH
E-value=0,
Score=4340
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis (PubMed:25558067). With evidence from 9 publications.
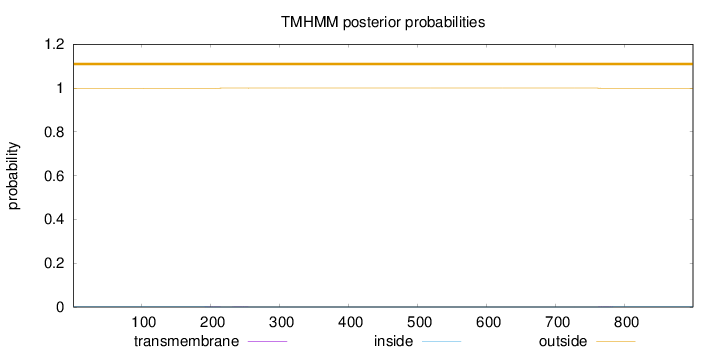
Length:
899
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03302
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00098
outside
1 - 899
Population Genetic Test Statistics
Pi
21.023039
Theta
20.67662
Tajima's D
-0.89538
CLR
0.874361
CSRT
0.157242137893105
Interpretation
Uncertain
