Gene
KWMTBOMO07271
Pre Gene Modal
BGIBMGA010601
Annotation
PREDICTED:_phospholipase_A1_member_A-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.609 PlasmaMembrane Reliability : 1.023
Sequence
CDS
ATGCAGTTCTTTCTTTACACCAGAACCACACAAGAAAAAGGTGAATTATTAAATACATTAGACAACAGATCTCTAACAGCCTCGAACTTCAATCCACTCCATCCAACGAAAATAGTCGTGCACGGATTCGGCGGAGGCAGGAACCTGTCGCCTAGTACTGATATGCGGAAGGCTTACTTCACAAGGGGCAATTACAACATCATCATCGTGGATTACGGAAGTCTCGTGAAAGAGCCGTGCTTAAGTCAGATAGAATGGGCTCCGCGTTTTGCCAGTCTCTGTATAACACAGCTGGTAGAATATTTACAATATCACCCAAAGAAGCAAGTTCAGCCGGAACAAATACATACTATTGGGTACAGTGTCGGCGCCCATATACTAGGCCTTGTGGCTAATTATTTAAAAGTCGGAAAACTTGGCAGGATAACTGGCTTAGACCCGACTATTTTCTTCTACATGGGCAACAATAGATCCCGTGACCTGGATCACACGGACGCGTTATTTGTGGACATTTTGCATACTGGAGCCGGAATCCTTGGCCAGTGGGGTCCGAACGGTCACGCGGATTTCTATGTCAACGGAGGCTCCAGTCAACCTGGTTGTGCTCATGATACCATATTCCAAACATTGTCCTGTGATCATACGAAGGTGACGCCGTACTTCATTGAGTCAATAAACAGCAGAGCCGGCTTCTGGGCGGGTCCTTGTTCTAGCTTGTTTTCATACTTAATAGGCTGGTGTGAACCCAGAGACACTGAATACGTGCTGATGGGAGAACATGTTAGTCATAATGCCAGAGGCGTATACTACGTCACCACTAACGCGAAGCCACCGTACGCTAGAGGCTTCCCGGGCAAATCGAGACGGCTTCGGCGACAGACATCACTCACCAAAAGCTGA
Protein
MQFFLYTRTTQEKGELLNTLDNRSLTASNFNPLHPTKIVVHGFGGGRNLSPSTDMRKAYFTRGNYNIIIVDYGSLVKEPCLSQIEWAPRFASLCITQLVEYLQYHPKKQVQPEQIHTIGYSVGAHILGLVANYLKVGKLGRITGLDPTIFFYMGNNRSRDLDHTDALFVDILHTGAGILGQWGPNGHADFYVNGGSSQPGCAHDTIFQTLSCDHTKVTPYFIESINSRAGFWAGPCSSLFSYLIGWCEPRDTEYVLMGEHVSHNARGVYYVTTNAKPPYARGFPGKSRRLRRQTSLTKS
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9JM49
A0A2A4IYM1
A0A2H1VSU8
A0A194R0T2
A0A212EWK3
A0A194PHF7
+ More
Q17JZ0 A0A182G951 A0A1W7R7I4 A0A182MGP8 A0A182RFJ2 A0A182F1K5 A0A182NTM1 A0A182QAJ5 A0A182P4R5 A0A182TH71 A0A1S4HCZ5 W5JX23 A0A182USW6 A0A182W7F0 A0A182IXY0 A0A182K5I9 A0A084WD64 A0A182WT18 A0A182HV15 A0A182XZX0 A0A1B0CT56 A0A336LVZ2 A0A1I8N877 A0A1A9VZW1 A0A1I8P8N2 A0A0L0C8B3 A0A1A9ZN25 A0A1B0A1K5 A0A1B0G6L5 A0A1A9V9G6 A0A1W4WWZ5 A0A1B0BGX8 D2A243 B4MTZ8 A0A0A1XNT0 A0A232ENH1 A0A3B0K7A0 W8BW47 B4I2K3 B3MLJ3 A0A1S4J177 A0A0J9QUB4 K7J3M6 A0A2J7PRY9 B3N9L3 Q9VQC2 A0A0J9QV40 B4KGU8 B4NWP4 E2AEA0 A0A1J1IQ41 A0A1W4V6J5 U4UIE9 F4X439 B4JAU7 E0W3V9 A0A151JX39 A0A067QXJ9 A0A0R3NU10 E2BJ46 B4GSK7 D0QWG9 Q29KZ1 A0A1B6L8G6 B4LUD6 A0A2S2NW92 A0A2H8TPN2 A0A2S2QRT7 A0A154NVX7 A0A3L8E4P5 A0A0L7LAL0 A0A088AT47 A0A151IIL5 A0A1B6J085 A0A1B6J9T3 A0A0C9R4X8 A0A146LBX8 A0A0N0BHI8 A0A0M4ENI1 A0A1B0DHZ7 A0A1W4WVX8 B0W0G0 A0A151HZU4 A0A151WS03 A0A151ISV1 B0W0G1 A0A1S4J0Q9 A0A0L7QV19 Q7Q3R1 T1H7M6 A0A1B6E4Y5
Q17JZ0 A0A182G951 A0A1W7R7I4 A0A182MGP8 A0A182RFJ2 A0A182F1K5 A0A182NTM1 A0A182QAJ5 A0A182P4R5 A0A182TH71 A0A1S4HCZ5 W5JX23 A0A182USW6 A0A182W7F0 A0A182IXY0 A0A182K5I9 A0A084WD64 A0A182WT18 A0A182HV15 A0A182XZX0 A0A1B0CT56 A0A336LVZ2 A0A1I8N877 A0A1A9VZW1 A0A1I8P8N2 A0A0L0C8B3 A0A1A9ZN25 A0A1B0A1K5 A0A1B0G6L5 A0A1A9V9G6 A0A1W4WWZ5 A0A1B0BGX8 D2A243 B4MTZ8 A0A0A1XNT0 A0A232ENH1 A0A3B0K7A0 W8BW47 B4I2K3 B3MLJ3 A0A1S4J177 A0A0J9QUB4 K7J3M6 A0A2J7PRY9 B3N9L3 Q9VQC2 A0A0J9QV40 B4KGU8 B4NWP4 E2AEA0 A0A1J1IQ41 A0A1W4V6J5 U4UIE9 F4X439 B4JAU7 E0W3V9 A0A151JX39 A0A067QXJ9 A0A0R3NU10 E2BJ46 B4GSK7 D0QWG9 Q29KZ1 A0A1B6L8G6 B4LUD6 A0A2S2NW92 A0A2H8TPN2 A0A2S2QRT7 A0A154NVX7 A0A3L8E4P5 A0A0L7LAL0 A0A088AT47 A0A151IIL5 A0A1B6J085 A0A1B6J9T3 A0A0C9R4X8 A0A146LBX8 A0A0N0BHI8 A0A0M4ENI1 A0A1B0DHZ7 A0A1W4WVX8 B0W0G0 A0A151HZU4 A0A151WS03 A0A151ISV1 B0W0G1 A0A1S4J0Q9 A0A0L7QV19 Q7Q3R1 T1H7M6 A0A1B6E4Y5
Pubmed
19121390
26354079
22118469
17510324
26483478
12364791
+ More
20920257 23761445 24438588 25244985 25315136 26108605 18362917 19820115 17994087 25830018 28648823 24495485 22936249 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 23537049 21719571 20566863 24845553 15632085 19859648 30249741 26227816 26823975
20920257 23761445 24438588 25244985 25315136 26108605 18362917 19820115 17994087 25830018 28648823 24495485 22936249 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20798317 23537049 21719571 20566863 24845553 15632085 19859648 30249741 26227816 26823975
EMBL
BABH01041851
NWSH01004567
PCG64915.1
ODYU01004240
SOQ43903.1
KQ460883
+ More
KPJ11119.1 AGBW02011969 OWR45847.1 KQ459603 KPI92846.1 CH477229 EAT47010.1 JXUM01048945 KQ561583 KXJ78100.1 GEHC01000527 JAV47118.1 AXCM01016815 AXCN02000795 AAAB01008964 ADMH02000054 ETN67975.1 ATLV01022945 ATLV01022946 ATLV01022947 KE525338 KFB48158.1 APCN01002489 AJWK01027122 AJWK01027123 UFQT01000238 SSX22222.1 JRES01000753 KNC28678.1 CCAG010023094 JXJN01013743 JXJN01014083 KQ971338 EFA01489.1 CH963852 EDW75587.2 GBXI01001636 JAD12656.1 NNAY01003153 OXU19900.1 OUUW01000006 SPP81899.1 GAMC01005387 JAC01169.1 CH480820 EDW53998.1 CH902620 EDV31742.2 CM002910 KMY87673.1 NEVH01021939 PNF19105.1 CH954177 EDV57470.2 AE014134 BT126300 AAF51254.3 AEB39661.1 KMY87674.1 CH933807 EDW11148.2 CM000157 EDW87386.2 GL438827 EFN68333.1 CVRI01000055 CRL01214.1 KB632373 ERL93779.1 GL888625 EGI58763.1 CH916368 EDW03905.1 DS235883 EEB20315.1 KQ981607 KYN39413.1 KK852854 KDR14960.1 CH379061 KRT04686.1 GL448558 EFN84196.1 CH479189 EDW25366.1 FJ821028 ACN94668.1 EAL33033.3 GEBQ01029841 GEBQ01019989 JAT10136.1 JAT19988.1 CH940649 EDW64122.2 GGMR01008703 MBY21322.1 GFXV01004300 MBW16105.1 GGMS01011252 MBY80455.1 KQ434771 KZC03826.1 QOIP01000001 RLU26948.1 JTDY01001928 KOB72528.1 KQ977477 KYN02492.1 GECU01015150 JAS92556.1 GECU01011774 JAS95932.1 GBYB01001896 JAG71663.1 GDHC01012848 JAQ05781.1 KQ435750 KOX76331.1 CP012523 ALC38225.1 AJVK01006113 DS231817 EDS39699.1 KQ976687 KYM78141.1 KQ982796 KYQ50598.1 KQ981064 KYN09847.1 EDS39700.1 KQ414731 KOC62399.1 EAA12397.4 ACPB03003282 ACPB03003283 ACPB03003284 GEDC01004334 JAS32964.1
KPJ11119.1 AGBW02011969 OWR45847.1 KQ459603 KPI92846.1 CH477229 EAT47010.1 JXUM01048945 KQ561583 KXJ78100.1 GEHC01000527 JAV47118.1 AXCM01016815 AXCN02000795 AAAB01008964 ADMH02000054 ETN67975.1 ATLV01022945 ATLV01022946 ATLV01022947 KE525338 KFB48158.1 APCN01002489 AJWK01027122 AJWK01027123 UFQT01000238 SSX22222.1 JRES01000753 KNC28678.1 CCAG010023094 JXJN01013743 JXJN01014083 KQ971338 EFA01489.1 CH963852 EDW75587.2 GBXI01001636 JAD12656.1 NNAY01003153 OXU19900.1 OUUW01000006 SPP81899.1 GAMC01005387 JAC01169.1 CH480820 EDW53998.1 CH902620 EDV31742.2 CM002910 KMY87673.1 NEVH01021939 PNF19105.1 CH954177 EDV57470.2 AE014134 BT126300 AAF51254.3 AEB39661.1 KMY87674.1 CH933807 EDW11148.2 CM000157 EDW87386.2 GL438827 EFN68333.1 CVRI01000055 CRL01214.1 KB632373 ERL93779.1 GL888625 EGI58763.1 CH916368 EDW03905.1 DS235883 EEB20315.1 KQ981607 KYN39413.1 KK852854 KDR14960.1 CH379061 KRT04686.1 GL448558 EFN84196.1 CH479189 EDW25366.1 FJ821028 ACN94668.1 EAL33033.3 GEBQ01029841 GEBQ01019989 JAT10136.1 JAT19988.1 CH940649 EDW64122.2 GGMR01008703 MBY21322.1 GFXV01004300 MBW16105.1 GGMS01011252 MBY80455.1 KQ434771 KZC03826.1 QOIP01000001 RLU26948.1 JTDY01001928 KOB72528.1 KQ977477 KYN02492.1 GECU01015150 JAS92556.1 GECU01011774 JAS95932.1 GBYB01001896 JAG71663.1 GDHC01012848 JAQ05781.1 KQ435750 KOX76331.1 CP012523 ALC38225.1 AJVK01006113 DS231817 EDS39699.1 KQ976687 KYM78141.1 KQ982796 KYQ50598.1 KQ981064 KYN09847.1 EDS39700.1 KQ414731 KOC62399.1 EAA12397.4 ACPB03003282 ACPB03003283 ACPB03003284 GEDC01004334 JAS32964.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000008820
+ More
UP000069940 UP000249989 UP000075883 UP000075900 UP000069272 UP000075884 UP000075886 UP000075885 UP000075902 UP000000673 UP000075903 UP000075920 UP000075880 UP000075881 UP000030765 UP000076407 UP000075840 UP000076408 UP000092461 UP000095301 UP000091820 UP000095300 UP000037069 UP000092445 UP000092444 UP000078200 UP000192223 UP000092460 UP000007266 UP000007798 UP000215335 UP000268350 UP000001292 UP000007801 UP000002358 UP000235965 UP000008711 UP000000803 UP000009192 UP000002282 UP000000311 UP000183832 UP000192221 UP000030742 UP000007755 UP000001070 UP000009046 UP000078541 UP000027135 UP000001819 UP000008237 UP000008744 UP000008792 UP000076502 UP000279307 UP000037510 UP000005203 UP000078542 UP000053105 UP000092553 UP000092462 UP000002320 UP000078540 UP000075809 UP000078492 UP000053825 UP000007062 UP000015103
UP000069940 UP000249989 UP000075883 UP000075900 UP000069272 UP000075884 UP000075886 UP000075885 UP000075902 UP000000673 UP000075903 UP000075920 UP000075880 UP000075881 UP000030765 UP000076407 UP000075840 UP000076408 UP000092461 UP000095301 UP000091820 UP000095300 UP000037069 UP000092445 UP000092444 UP000078200 UP000192223 UP000092460 UP000007266 UP000007798 UP000215335 UP000268350 UP000001292 UP000007801 UP000002358 UP000235965 UP000008711 UP000000803 UP000009192 UP000002282 UP000000311 UP000183832 UP000192221 UP000030742 UP000007755 UP000001070 UP000009046 UP000078541 UP000027135 UP000001819 UP000008237 UP000008744 UP000008792 UP000076502 UP000279307 UP000037510 UP000005203 UP000078542 UP000053105 UP000092553 UP000092462 UP000002320 UP000078540 UP000075809 UP000078492 UP000053825 UP000007062 UP000015103
Interpro
Gene 3D
ProteinModelPortal
H9JM49
A0A2A4IYM1
A0A2H1VSU8
A0A194R0T2
A0A212EWK3
A0A194PHF7
+ More
Q17JZ0 A0A182G951 A0A1W7R7I4 A0A182MGP8 A0A182RFJ2 A0A182F1K5 A0A182NTM1 A0A182QAJ5 A0A182P4R5 A0A182TH71 A0A1S4HCZ5 W5JX23 A0A182USW6 A0A182W7F0 A0A182IXY0 A0A182K5I9 A0A084WD64 A0A182WT18 A0A182HV15 A0A182XZX0 A0A1B0CT56 A0A336LVZ2 A0A1I8N877 A0A1A9VZW1 A0A1I8P8N2 A0A0L0C8B3 A0A1A9ZN25 A0A1B0A1K5 A0A1B0G6L5 A0A1A9V9G6 A0A1W4WWZ5 A0A1B0BGX8 D2A243 B4MTZ8 A0A0A1XNT0 A0A232ENH1 A0A3B0K7A0 W8BW47 B4I2K3 B3MLJ3 A0A1S4J177 A0A0J9QUB4 K7J3M6 A0A2J7PRY9 B3N9L3 Q9VQC2 A0A0J9QV40 B4KGU8 B4NWP4 E2AEA0 A0A1J1IQ41 A0A1W4V6J5 U4UIE9 F4X439 B4JAU7 E0W3V9 A0A151JX39 A0A067QXJ9 A0A0R3NU10 E2BJ46 B4GSK7 D0QWG9 Q29KZ1 A0A1B6L8G6 B4LUD6 A0A2S2NW92 A0A2H8TPN2 A0A2S2QRT7 A0A154NVX7 A0A3L8E4P5 A0A0L7LAL0 A0A088AT47 A0A151IIL5 A0A1B6J085 A0A1B6J9T3 A0A0C9R4X8 A0A146LBX8 A0A0N0BHI8 A0A0M4ENI1 A0A1B0DHZ7 A0A1W4WVX8 B0W0G0 A0A151HZU4 A0A151WS03 A0A151ISV1 B0W0G1 A0A1S4J0Q9 A0A0L7QV19 Q7Q3R1 T1H7M6 A0A1B6E4Y5
Q17JZ0 A0A182G951 A0A1W7R7I4 A0A182MGP8 A0A182RFJ2 A0A182F1K5 A0A182NTM1 A0A182QAJ5 A0A182P4R5 A0A182TH71 A0A1S4HCZ5 W5JX23 A0A182USW6 A0A182W7F0 A0A182IXY0 A0A182K5I9 A0A084WD64 A0A182WT18 A0A182HV15 A0A182XZX0 A0A1B0CT56 A0A336LVZ2 A0A1I8N877 A0A1A9VZW1 A0A1I8P8N2 A0A0L0C8B3 A0A1A9ZN25 A0A1B0A1K5 A0A1B0G6L5 A0A1A9V9G6 A0A1W4WWZ5 A0A1B0BGX8 D2A243 B4MTZ8 A0A0A1XNT0 A0A232ENH1 A0A3B0K7A0 W8BW47 B4I2K3 B3MLJ3 A0A1S4J177 A0A0J9QUB4 K7J3M6 A0A2J7PRY9 B3N9L3 Q9VQC2 A0A0J9QV40 B4KGU8 B4NWP4 E2AEA0 A0A1J1IQ41 A0A1W4V6J5 U4UIE9 F4X439 B4JAU7 E0W3V9 A0A151JX39 A0A067QXJ9 A0A0R3NU10 E2BJ46 B4GSK7 D0QWG9 Q29KZ1 A0A1B6L8G6 B4LUD6 A0A2S2NW92 A0A2H8TPN2 A0A2S2QRT7 A0A154NVX7 A0A3L8E4P5 A0A0L7LAL0 A0A088AT47 A0A151IIL5 A0A1B6J085 A0A1B6J9T3 A0A0C9R4X8 A0A146LBX8 A0A0N0BHI8 A0A0M4ENI1 A0A1B0DHZ7 A0A1W4WVX8 B0W0G0 A0A151HZU4 A0A151WS03 A0A151ISV1 B0W0G1 A0A1S4J0Q9 A0A0L7QV19 Q7Q3R1 T1H7M6 A0A1B6E4Y5
PDB
1GPL
E-value=1.22166e-27,
Score=305
Ontologies
GO
PANTHER
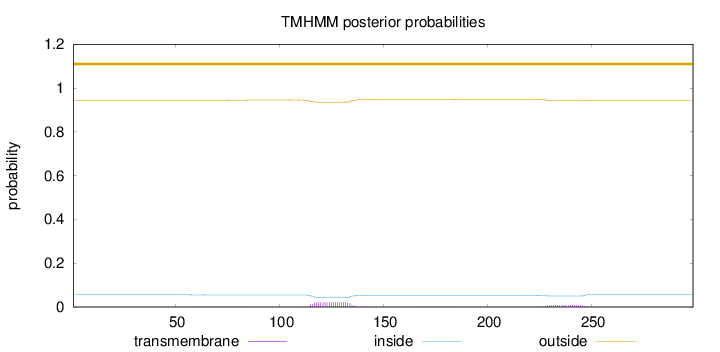
Topology
Subcellular location
Secreted
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.60661
Exp number, first 60 AAs:
0.00128
Total prob of N-in:
0.05546
outside
1 - 299
Population Genetic Test Statistics
Pi
5.608959
Theta
14.911761
Tajima's D
-1.574226
CLR
18.444353
CSRT
0.0515974201289935
Interpretation
Uncertain
