Gene
KWMTBOMO07270
Pre Gene Modal
BGIBMGA010600
Annotation
PREDICTED:_pancreatic_lipase-related_protein_3-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.718
Sequence
CDS
ATGTTACGACGTCTATTATACTCTCATGGATTTTTTCTCTGTATAATAACAGCGTGCTTAAGTTGTGTTACGCCACCTTTGGAATGTCCAAATAAAAATATTACTTTCTGGTTGTACACTCGGGCCAACGAACACAGTCCTCATCAACTATTAGCCGATGACAACAATTCTATAACCGAAGCACCGTGGGTTACTGACGCTCCGATCAAGATACTGATCCACGGGTACACGGGTCATAGAGATTTCTCACCTAACACTGAAATTAGACCTGCCTACATGAAATGTTGCGATTACAATGTTATTAGTGTGGACTATAATAACATAGCATTGGAGCCTTGCTATTTGGAATCAGCTAGAAACACAGAGCTAGTTGGAAAATGTACCGCTCAACTTATAGATAATCTGATCAAAATACATGGTTTTAAGATGCAAAGATTTCACGTGATCGGATTCAGTCTTGGTGCTCAAACAGCCGGTTACATGGCAAATTACCTAATCAATGGACGACTATCGAGAATTACCGCATTAGATCCTGCTATGCCCCTGTTTGTTACAACTGACAACAGTAGAAAGGTTGACATGACTGATGCAGATTTTGTAGACGTTCTCCATACGAATGCTTTAGAAAAAGGAAAATTAGAAGCCAGCGGTCATGTAGACTTTTACGCCAACGGTGGTCTCACTCAACCCGGCTGTAAAGCTTCTGAAGAACAAACAAAATCCGGTTGCGACCACGCCAGGGCACCGATATACTACGCGGAGTCTATTATAACAGACGTTGGTTTTTACGCAACGAAATGTTTTTCTTGGATCACTTACATGATCGGCTGGTGTGAGCTAATCAATTCGAGTGATGAGATATTATTTGGCGAATATGTATCTTTGGAAGTCACCGGTTTTTATTTCTTCCCGACAAATTTTGAATCTCCATTCGCCAAAGGTTTGAACCACAGGAACACAAGAGATTTATTAAGTCAAGAATTTAATAAATATGATAATGGAATTTGA
Protein
MLRRLLYSHGFFLCIITACLSCVTPPLECPNKNITFWLYTRANEHSPHQLLADDNNSITEAPWVTDAPIKILIHGYTGHRDFSPNTEIRPAYMKCCDYNVISVDYNNIALEPCYLESARNTELVGKCTAQLIDNLIKIHGFKMQRFHVIGFSLGAQTAGYMANYLINGRLSRITALDPAMPLFVTTDNSRKVDMTDADFVDVLHTNALEKGKLEASGHVDFYANGGLTQPGCKASEEQTKSGCDHARAPIYYAESIITDVGFYATKCFSWITYMIGWCELINSSDEILFGEYVSLEVTGFYFFPTNFESPFAKGLNHRNTRDLLSQEFNKYDNGI
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A2H1VTT1
A0A2A4IZK1
A0A212EWK3
A0A194PNZ2
A0A0L7LAL0
A0A194R1K0
+ More
A0A1S4JCE7 Q17JZ1 A0A1S4EZX5 A0A182P4R3 A0A182JWK1 A0A1S4GWY0 A0A182L1W4 A0A182NTL9 A0A182VCY9 A0A182JFQ9 A0A182QH31 A0A182F1K3 A0A182W7F1 A0A336KAB5 B0WCH8 A0A182TJ83 A0A182G950 A0A182XZX0 W5JV14 A0A182RFJ3 A0A182S7Y1 A0A182HV15 W8C3L2 A0A182LVZ5 A0A3B0JHS5 B4LUD8 B4JAU5 A0A3B0K8D7 B3N9L6 A0A1B0DHZ8 B4KGU7 J9JMV1 A0A1J1IQ41 A0A0R1DIS3 B4MU00 B5DJJ6 Q4V566 D0QWH0 A0A1W4UT91 B4I2K6 B4GSK8 Q4V563 B4Q844 Q7Q3R1 A0A1I8PI98 A0A0M4E6L6 A0A1I8MEV2 A0A1B6C008 A0A0A1WTW2 A0A0M5IZ10 A0A0Q5WL87 A0A0M3QTX4 A0A0L0C8C6 Q17JZ0 A0A182G951 A0A2A4IYM1 A0A232ENH1 A0A1W7R7I4 A0A0J7KTU3 B4P084 A0A0L7REF5 E0W3W0 K7J3M6 A0A310SBJ7 A0A0P8XG42 A0A154NZG7 A0A2H1VSU8 A0A1B0CT56 A0A182F1K5 E9IHA8 E2ATG3 A0A1I9WLB3 Q0E8T1 Q4V5V9 F4WV93 E2AEA0 A0A151JX39 Q3HKQ5 B4I1E2 F4X439 A0A182P4R5 A0A1S4HCZ5 A0A0J9QW48 B4Q3J7 A0A1B0G6L5 A0A1I8N877 A0A154NVX7 A0A1B0A1K5 A0A1A9ZN25 A0A182IXY0
A0A1S4JCE7 Q17JZ1 A0A1S4EZX5 A0A182P4R3 A0A182JWK1 A0A1S4GWY0 A0A182L1W4 A0A182NTL9 A0A182VCY9 A0A182JFQ9 A0A182QH31 A0A182F1K3 A0A182W7F1 A0A336KAB5 B0WCH8 A0A182TJ83 A0A182G950 A0A182XZX0 W5JV14 A0A182RFJ3 A0A182S7Y1 A0A182HV15 W8C3L2 A0A182LVZ5 A0A3B0JHS5 B4LUD8 B4JAU5 A0A3B0K8D7 B3N9L6 A0A1B0DHZ8 B4KGU7 J9JMV1 A0A1J1IQ41 A0A0R1DIS3 B4MU00 B5DJJ6 Q4V566 D0QWH0 A0A1W4UT91 B4I2K6 B4GSK8 Q4V563 B4Q844 Q7Q3R1 A0A1I8PI98 A0A0M4E6L6 A0A1I8MEV2 A0A1B6C008 A0A0A1WTW2 A0A0M5IZ10 A0A0Q5WL87 A0A0M3QTX4 A0A0L0C8C6 Q17JZ0 A0A182G951 A0A2A4IYM1 A0A232ENH1 A0A1W7R7I4 A0A0J7KTU3 B4P084 A0A0L7REF5 E0W3W0 K7J3M6 A0A310SBJ7 A0A0P8XG42 A0A154NZG7 A0A2H1VSU8 A0A1B0CT56 A0A182F1K5 E9IHA8 E2ATG3 A0A1I9WLB3 Q0E8T1 Q4V5V9 F4WV93 E2AEA0 A0A151JX39 Q3HKQ5 B4I1E2 F4X439 A0A182P4R5 A0A1S4HCZ5 A0A0J9QW48 B4Q3J7 A0A1B0G6L5 A0A1I8N877 A0A154NVX7 A0A1B0A1K5 A0A1A9ZN25 A0A182IXY0
Pubmed
22118469
26354079
26227816
17510324
12364791
20966253
+ More
26483478 25244985 20920257 23761445 24495485 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 19859648 22936249 25315136 25830018 26108605 28648823 20566863 20075255 21282665 20798317 27538518 21719571 15944345
26483478 25244985 20920257 23761445 24495485 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 19859648 22936249 25315136 25830018 26108605 28648823 20566863 20075255 21282665 20798317 27538518 21719571 15944345
EMBL
ODYU01004240
SOQ43902.1
NWSH01004567
PCG64916.1
AGBW02011969
OWR45847.1
+ More
KQ459603 KPI92845.1 JTDY01001928 KOB72528.1 KQ460883 KPJ11120.1 CH477229 EAT47009.1 AAAB01008964 AXCN02000795 UFQS01000238 UFQT01000238 SSX01844.1 SSX22221.1 DS231888 EDS43549.1 JXUM01048933 JXUM01048934 KQ561583 KXJ78099.1 ADMH02000054 ETN67976.1 APCN01002489 GAMC01000013 JAC06543.1 AXCM01009280 OUUW01000006 SPP81897.1 CH940649 EDW64124.1 CH916368 EDW03903.1 SPP81896.1 CH954177 EDV57473.2 AJVK01006113 AJVK01006114 CH933807 EDW11147.1 ABLF02034706 ABLF02034708 CVRI01000055 CRL01214.1 CM000157 KRJ97140.1 CH963852 EDW75589.1 CH379061 EDY70468.1 BT022790 AE014134 AAY55206.1 ABV53607.1 FJ821028 ACN94669.1 CH480820 EDW54001.1 CH479189 EDW25367.1 BT022793 AAY55209.1 CM000361 CM002910 EDX03466.1 KMY87677.1 EAA12397.4 CP012523 ALC38227.1 GEDC01030460 JAS06838.1 GBXI01012464 JAD01828.1 ALC39636.1 ALC39641.1 KQS70175.1 ALC39639.1 JRES01000753 KNC28693.1 EAT47010.1 JXUM01048945 KXJ78100.1 PCG64915.1 NNAY01003153 OXU19900.1 GEHC01000527 JAV47118.1 LBMM01003255 KMQ93748.1 EDW88949.2 KQ414611 KOC69214.1 DS235883 EEB20316.1 KQ770887 OAD52563.1 CH902620 KPU73605.1 KQ434777 KZC04270.1 SOQ43903.1 AJWK01027122 AJWK01027123 GL763196 EFZ20041.1 GL442566 EFN63276.1 KU932297 APA33933.1 ABI31293.1 AHN54133.1 BT022547 AAY54963.1 GL888384 EGI61859.1 GL438827 EFN68333.1 KQ981607 KYN39413.1 DQ231603 ABA71709.1 EDW54349.1 GL888625 EGI58763.1 KMY88266.1 EDX03801.1 CCAG010023094 KQ434771 KZC03826.1
KQ459603 KPI92845.1 JTDY01001928 KOB72528.1 KQ460883 KPJ11120.1 CH477229 EAT47009.1 AAAB01008964 AXCN02000795 UFQS01000238 UFQT01000238 SSX01844.1 SSX22221.1 DS231888 EDS43549.1 JXUM01048933 JXUM01048934 KQ561583 KXJ78099.1 ADMH02000054 ETN67976.1 APCN01002489 GAMC01000013 JAC06543.1 AXCM01009280 OUUW01000006 SPP81897.1 CH940649 EDW64124.1 CH916368 EDW03903.1 SPP81896.1 CH954177 EDV57473.2 AJVK01006113 AJVK01006114 CH933807 EDW11147.1 ABLF02034706 ABLF02034708 CVRI01000055 CRL01214.1 CM000157 KRJ97140.1 CH963852 EDW75589.1 CH379061 EDY70468.1 BT022790 AE014134 AAY55206.1 ABV53607.1 FJ821028 ACN94669.1 CH480820 EDW54001.1 CH479189 EDW25367.1 BT022793 AAY55209.1 CM000361 CM002910 EDX03466.1 KMY87677.1 EAA12397.4 CP012523 ALC38227.1 GEDC01030460 JAS06838.1 GBXI01012464 JAD01828.1 ALC39636.1 ALC39641.1 KQS70175.1 ALC39639.1 JRES01000753 KNC28693.1 EAT47010.1 JXUM01048945 KXJ78100.1 PCG64915.1 NNAY01003153 OXU19900.1 GEHC01000527 JAV47118.1 LBMM01003255 KMQ93748.1 EDW88949.2 KQ414611 KOC69214.1 DS235883 EEB20316.1 KQ770887 OAD52563.1 CH902620 KPU73605.1 KQ434777 KZC04270.1 SOQ43903.1 AJWK01027122 AJWK01027123 GL763196 EFZ20041.1 GL442566 EFN63276.1 KU932297 APA33933.1 ABI31293.1 AHN54133.1 BT022547 AAY54963.1 GL888384 EGI61859.1 GL438827 EFN68333.1 KQ981607 KYN39413.1 DQ231603 ABA71709.1 EDW54349.1 GL888625 EGI58763.1 KMY88266.1 EDX03801.1 CCAG010023094 KQ434771 KZC03826.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000037510
UP000053240
UP000008820
+ More
UP000075885 UP000075881 UP000075882 UP000075884 UP000075903 UP000075880 UP000075886 UP000069272 UP000075920 UP000002320 UP000075902 UP000069940 UP000249989 UP000076408 UP000000673 UP000075900 UP000075901 UP000075840 UP000075883 UP000268350 UP000008792 UP000001070 UP000008711 UP000092462 UP000009192 UP000007819 UP000183832 UP000002282 UP000007798 UP000001819 UP000000803 UP000192221 UP000001292 UP000008744 UP000000304 UP000007062 UP000095300 UP000092553 UP000095301 UP000037069 UP000215335 UP000036403 UP000053825 UP000009046 UP000002358 UP000007801 UP000076502 UP000092461 UP000000311 UP000007755 UP000078541 UP000092444 UP000092445
UP000075885 UP000075881 UP000075882 UP000075884 UP000075903 UP000075880 UP000075886 UP000069272 UP000075920 UP000002320 UP000075902 UP000069940 UP000249989 UP000076408 UP000000673 UP000075900 UP000075901 UP000075840 UP000075883 UP000268350 UP000008792 UP000001070 UP000008711 UP000092462 UP000009192 UP000007819 UP000183832 UP000002282 UP000007798 UP000001819 UP000000803 UP000192221 UP000001292 UP000008744 UP000000304 UP000007062 UP000095300 UP000092553 UP000095301 UP000037069 UP000215335 UP000036403 UP000053825 UP000009046 UP000002358 UP000007801 UP000076502 UP000092461 UP000000311 UP000007755 UP000078541 UP000092444 UP000092445
Interpro
IPR013818
Lipase/vitellogenin
+ More
IPR029058 AB_hydrolase
IPR033906 Lipase_N
IPR000734 TAG_lipase
IPR013032 EGF-like_CS
IPR001791 Laminin_G
IPR000742 EGF-like_dom
IPR001881 EGF-like_Ca-bd_dom
IPR013320 ConA-like_dom_sf
IPR016272 Lipase_LIPH
IPR023415 LDLR_class-A_CS
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR029058 AB_hydrolase
IPR033906 Lipase_N
IPR000734 TAG_lipase
IPR013032 EGF-like_CS
IPR001791 Laminin_G
IPR000742 EGF-like_dom
IPR001881 EGF-like_Ca-bd_dom
IPR013320 ConA-like_dom_sf
IPR016272 Lipase_LIPH
IPR023415 LDLR_class-A_CS
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
Gene 3D
ProteinModelPortal
A0A2H1VTT1
A0A2A4IZK1
A0A212EWK3
A0A194PNZ2
A0A0L7LAL0
A0A194R1K0
+ More
A0A1S4JCE7 Q17JZ1 A0A1S4EZX5 A0A182P4R3 A0A182JWK1 A0A1S4GWY0 A0A182L1W4 A0A182NTL9 A0A182VCY9 A0A182JFQ9 A0A182QH31 A0A182F1K3 A0A182W7F1 A0A336KAB5 B0WCH8 A0A182TJ83 A0A182G950 A0A182XZX0 W5JV14 A0A182RFJ3 A0A182S7Y1 A0A182HV15 W8C3L2 A0A182LVZ5 A0A3B0JHS5 B4LUD8 B4JAU5 A0A3B0K8D7 B3N9L6 A0A1B0DHZ8 B4KGU7 J9JMV1 A0A1J1IQ41 A0A0R1DIS3 B4MU00 B5DJJ6 Q4V566 D0QWH0 A0A1W4UT91 B4I2K6 B4GSK8 Q4V563 B4Q844 Q7Q3R1 A0A1I8PI98 A0A0M4E6L6 A0A1I8MEV2 A0A1B6C008 A0A0A1WTW2 A0A0M5IZ10 A0A0Q5WL87 A0A0M3QTX4 A0A0L0C8C6 Q17JZ0 A0A182G951 A0A2A4IYM1 A0A232ENH1 A0A1W7R7I4 A0A0J7KTU3 B4P084 A0A0L7REF5 E0W3W0 K7J3M6 A0A310SBJ7 A0A0P8XG42 A0A154NZG7 A0A2H1VSU8 A0A1B0CT56 A0A182F1K5 E9IHA8 E2ATG3 A0A1I9WLB3 Q0E8T1 Q4V5V9 F4WV93 E2AEA0 A0A151JX39 Q3HKQ5 B4I1E2 F4X439 A0A182P4R5 A0A1S4HCZ5 A0A0J9QW48 B4Q3J7 A0A1B0G6L5 A0A1I8N877 A0A154NVX7 A0A1B0A1K5 A0A1A9ZN25 A0A182IXY0
A0A1S4JCE7 Q17JZ1 A0A1S4EZX5 A0A182P4R3 A0A182JWK1 A0A1S4GWY0 A0A182L1W4 A0A182NTL9 A0A182VCY9 A0A182JFQ9 A0A182QH31 A0A182F1K3 A0A182W7F1 A0A336KAB5 B0WCH8 A0A182TJ83 A0A182G950 A0A182XZX0 W5JV14 A0A182RFJ3 A0A182S7Y1 A0A182HV15 W8C3L2 A0A182LVZ5 A0A3B0JHS5 B4LUD8 B4JAU5 A0A3B0K8D7 B3N9L6 A0A1B0DHZ8 B4KGU7 J9JMV1 A0A1J1IQ41 A0A0R1DIS3 B4MU00 B5DJJ6 Q4V566 D0QWH0 A0A1W4UT91 B4I2K6 B4GSK8 Q4V563 B4Q844 Q7Q3R1 A0A1I8PI98 A0A0M4E6L6 A0A1I8MEV2 A0A1B6C008 A0A0A1WTW2 A0A0M5IZ10 A0A0Q5WL87 A0A0M3QTX4 A0A0L0C8C6 Q17JZ0 A0A182G951 A0A2A4IYM1 A0A232ENH1 A0A1W7R7I4 A0A0J7KTU3 B4P084 A0A0L7REF5 E0W3W0 K7J3M6 A0A310SBJ7 A0A0P8XG42 A0A154NZG7 A0A2H1VSU8 A0A1B0CT56 A0A182F1K5 E9IHA8 E2ATG3 A0A1I9WLB3 Q0E8T1 Q4V5V9 F4WV93 E2AEA0 A0A151JX39 Q3HKQ5 B4I1E2 F4X439 A0A182P4R5 A0A1S4HCZ5 A0A0J9QW48 B4Q3J7 A0A1B0G6L5 A0A1I8N877 A0A154NVX7 A0A1B0A1K5 A0A1A9ZN25 A0A182IXY0
PDB
1W52
E-value=2.44634e-30,
Score=329
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
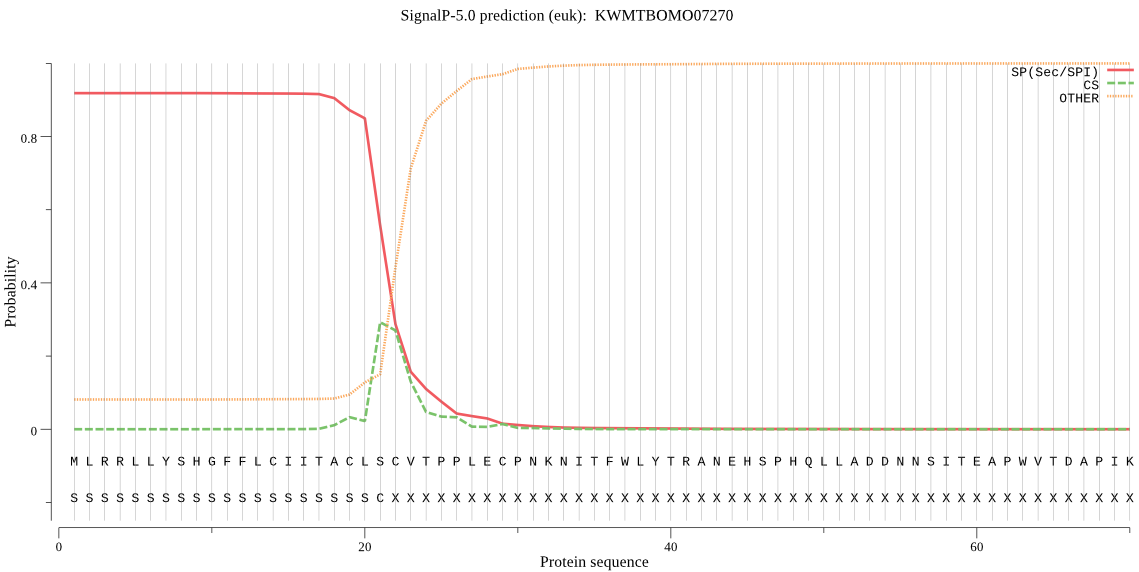
SignalP
Position: 1 - 21,
Likelihood: 0.918743
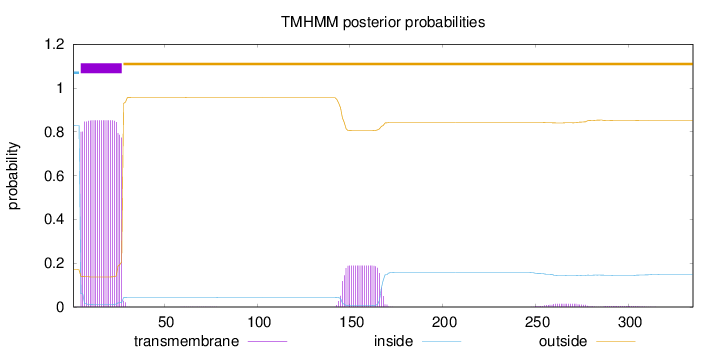
Length:
335
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.80845
Exp number, first 60 AAs:
19.33176
Total prob of N-in:
0.82953
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 335
Population Genetic Test Statistics
Pi
15.205757
Theta
17.485634
Tajima's D
-1.316841
CLR
0.584195
CSRT
0.0806459677016149
Interpretation
Uncertain
