Gene
KWMTBOMO07268
Pre Gene Modal
BGIBMGA010597
Annotation
PREDICTED:_cadherin-related_tumor_suppressor-like?_partial_[Bombyx_mori]
Full name
Cadherin-related tumor suppressor
Alternative Name
Protein fat
Location in the cell
Extracellular Reliability : 0.978
Sequence
CDS
ATGACGAGGATGTGGCGATGGCACGTGGCGCTGGCGGTGCTGGCGCTGCTGGGTGGCGCAGGCGTGACCGCCGTCGCCGAGCAGATGCAGTCGCGCGCCGCCGACACCGGTGTCGATCTTCGAGTCCCCGAGGATCAACCGATCGGTACTCTTGTCGGTAGGATTCCAATTAAGCCTGGCTTCACGTACCGTTTTAATGAGCCTCCCAAAGAATTTGTTCTCGACCCTGTATCTGGCGAGATAAGGACGAATGTAGTGCTGGACCGGGAAACTGTTGATCGCTATGCGTTCGTGGTGCTCTCAAGTCAGCCCACGTATCCTATAGAAGTGCGATTAAGGGTAACGGATGTAAACGATAATTACCCAGAGTTCCCGGAACCTAGTATCGGTGTGACGTTATCGGAAAGTACAGCTGCGGGAACTAAGCTTCTTCTCGATGCAGCGACTGATAAGGATTTAGGTGAGAATGGTATCACTAATGACTACAGAATAATAGACGGCGACACTGAAGGGAAATTTAGACTGAACGTTACCGTTAATCCGAGCGGTCAGACTTCTTATTTGCATTTAGAAACTACAGGAAAACTAGACAGAGAAACAACAGATTTTTACGTGTTAAACATCTCTGCCCGCGATGGCGGTAATCCTCCGAAATTTGGATACCTTCAAGTGAATGTTAGCATATTGGATGTAAACGACAACCCGCCTATATTCGACCAAAGTGATTTCTCTGTGTCGTTGAATGAGAGCGTGCCTCCTGGAACCACTGTCTTAAAAGTAACAGCCACTGATAATGATTTAGGAGACAATAGTAAAATTACGTACGAGGTCATGGACACAGAAAAACAGTTCGCCGTAGATCCTGAATCGGGCGTGATAACAACTAGTAGAATATTAAACTGTCCTAAATATTGTAGTAATGCTACATGTAACATGACATGTGTATTGACAGTCATAGCTAAAGATCACGGTGTACCACGACAAGATGCCCGGACTTATTCGATGAAAATGCCAAGAATGGGTCACTTGTTGCCGCCATCACTGTTACTGATTTGGATGCCGGCTTGA
Protein
MTRMWRWHVALAVLALLGGAGVTAVAEQMQSRAADTGVDLRVPEDQPIGTLVGRIPIKPGFTYRFNEPPKEFVLDPVSGEIRTNVVLDRETVDRYAFVVLSSQPTYPIEVRLRVTDVNDNYPEFPEPSIGVTLSESTAAGTKLLLDAATDKDLGENGITNDYRIIDGDTEGKFRLNVTVNPSGQTSYLHLETTGKLDRETTDFYVLNISARDGGNPPKFGYLQVNVSILDVNDNPPIFDQSDFSVSLNESVPPGTTVLKVTATDNDLGDNSKITYEVMDTEKQFAVDPESGVITTSRILNCPKYCSNATCNMTCVLTVIAKDHGVPRQDARTYSMKMPRMGHLLPPSLLLIWMPA
Summary
Description
Involved in regulation of planar cell polarity in the compound eye where it is required for correct specification of the R3 and R4 photoreceptor cells by regulating Fz activity in the R3/R4 precursor cells (PubMed:11893338). This is likely to occur through creation of an ft gradient so that the equatorial R3/R4 precursor cell has a higher level of ft function than its polar neighbor (PubMed:15548581). Also required for planar cell polarity of wing hairs (PubMed:12540853, PubMed:15240556). Mediates heterophilic cell adhesion in vitro and is required to stabilize ds on the cell surface (PubMed:15240556). Involved in regulation of eye imaginal disk size (PubMed:23667559). Upstream component of the Hippo pathway where it is likely to act as a cell surface receptor involved in regulation of tissue size and is required for the localization and stability of ex (PubMed:16996265). Probably acts as a cell surface receptor for ds (PubMed:20434335).
Ft-mito: Regulates mitochondrial electron transport chain integrity and promotes oxidative phosphorylation.
Ft-mito: Regulates mitochondrial electron transport chain integrity and promotes oxidative phosphorylation.
Subunit
Interacts with Fbxl7 (PubMed:25107277). Ft-mito interacts with NADH dehydrogenase subunit ND-24 and with ATP synthase subunit ATPsynC (PubMed:25215488).
Miscellaneous
The name 'fat' originates from weak mutant alleles that exhibit a broadening of the abdomen.
Keywords
Calcium
Cell adhesion
Cell membrane
Complete proteome
Disulfide bond
EGF-like domain
Glycoprotein
Membrane
Mitochondrion
Phosphoprotein
Receptor
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Cadherin-related tumor suppressor
Uniprot
H9JM45
A0A2A4JGB0
A0A194R096
A0A0L7LEK0
A0A212EWJ7
A0A194PIG7
+ More
A0A1B0BXS9 A0A139WIU3 A0A1W4WS50 A0A2P8XK47 U4UMJ9 A0A0K8WIK8 A0A0K8WGR0 A0A1J1ILU1 A0A1B0D4J3 W8BM12 A0A182GBS5 W8BXI0 A0A0L0C4R4 B0W5P9 A0A1A9W0F1 A0A1A9XH88 A0A067RK74 A0A1A9UMV7 A0A1B0BXS8 A0A1A9ZJ37 B4N0N1 A0A182JZ92 A0A182PJ70 N6UFF5 A0A1B0FNU6 A0A182MKD8 A0A182F4L6 A0A182UAH5 A0A0M4E2E8 A0A182W7V5 A0A182VN95 A0A182WT87 B4I369 A0A182HV80 A0A182RBM1 A0A182SG56 W5J9S7 A0A182Q423 P33450 A0A0J9QVG6 A0A182YGU6 B3N3P8 A0A182IV90 A0A2R7WSJ6 A0A1W4UQG6 A0A1S4GXD7 A0A3B0JEL3 Q7Q3K5 B3MLP3 B4NYI2 Q29NP3 T1I617 A0A1I8MY28 A0A1I8P2A4 B4MDC5 A0A0M8ZZC1 E9IY59 B4JD41 A0A2A3ENR6 A0A2A3EQA1 A0A195D170 A0A0L7RBN5 E1ZWJ5 A0A088A7Z0 A0A195ERJ8 E2C5B8 E9HFS0 A0A0J7NXR0 A0A3L8DKB9 F4WQR0 A0A195BRJ0 A0A0P5CCQ8 A0A158NRY6 A0A0N8D072 A0A0P6J127 A0A0P5ZTU5 A0A0P4YMD5 A0A0P5B7A8 A0A0P5B1P8 A0A0P4YB10 A0A0P4XXI2 A0A0P5U2L5 A0A0P4ZCX0 A0A0N7ZFF4 A0A0P4YSZ5 A0A195D885 A0A151X3F5 E0VP03 K7ILV2 A0A3Q0JFL9 Q0IF47 A0A182L245 B7P9Y6 A0A1D2MXS1
A0A1B0BXS9 A0A139WIU3 A0A1W4WS50 A0A2P8XK47 U4UMJ9 A0A0K8WIK8 A0A0K8WGR0 A0A1J1ILU1 A0A1B0D4J3 W8BM12 A0A182GBS5 W8BXI0 A0A0L0C4R4 B0W5P9 A0A1A9W0F1 A0A1A9XH88 A0A067RK74 A0A1A9UMV7 A0A1B0BXS8 A0A1A9ZJ37 B4N0N1 A0A182JZ92 A0A182PJ70 N6UFF5 A0A1B0FNU6 A0A182MKD8 A0A182F4L6 A0A182UAH5 A0A0M4E2E8 A0A182W7V5 A0A182VN95 A0A182WT87 B4I369 A0A182HV80 A0A182RBM1 A0A182SG56 W5J9S7 A0A182Q423 P33450 A0A0J9QVG6 A0A182YGU6 B3N3P8 A0A182IV90 A0A2R7WSJ6 A0A1W4UQG6 A0A1S4GXD7 A0A3B0JEL3 Q7Q3K5 B3MLP3 B4NYI2 Q29NP3 T1I617 A0A1I8MY28 A0A1I8P2A4 B4MDC5 A0A0M8ZZC1 E9IY59 B4JD41 A0A2A3ENR6 A0A2A3EQA1 A0A195D170 A0A0L7RBN5 E1ZWJ5 A0A088A7Z0 A0A195ERJ8 E2C5B8 E9HFS0 A0A0J7NXR0 A0A3L8DKB9 F4WQR0 A0A195BRJ0 A0A0P5CCQ8 A0A158NRY6 A0A0N8D072 A0A0P6J127 A0A0P5ZTU5 A0A0P4YMD5 A0A0P5B7A8 A0A0P5B1P8 A0A0P4YB10 A0A0P4XXI2 A0A0P5U2L5 A0A0P4ZCX0 A0A0N7ZFF4 A0A0P4YSZ5 A0A195D885 A0A151X3F5 E0VP03 K7ILV2 A0A3Q0JFL9 Q0IF47 A0A182L245 B7P9Y6 A0A1D2MXS1
Pubmed
19121390
26354079
26227816
22118469
18362917
19820115
+ More
29403074 23537049 24495485 26483478 26108605 24845553 17994087 20920257 23761445 1959133 10731132 12537572 11893338 12540853 15240556 15548581 16687445 16996265 18327897 18635802 19574458 20434335 23667559 25215488 25107277 22936249 25244985 12364791 17550304 15632085 23185243 25315136 21282665 20798317 21292972 30249741 21719571 21347285 20566863 20075255 17510324 20966253 27289101
29403074 23537049 24495485 26483478 26108605 24845553 17994087 20920257 23761445 1959133 10731132 12537572 11893338 12540853 15240556 15548581 16687445 16996265 18327897 18635802 19574458 20434335 23667559 25215488 25107277 22936249 25244985 12364791 17550304 15632085 23185243 25315136 21282665 20798317 21292972 30249741 21719571 21347285 20566863 20075255 17510324 20966253 27289101
EMBL
BABH01039937
BABH01039938
NWSH01001485
PCG71107.1
KQ460883
KPJ11122.1
+ More
JTDY01001533 KOB73626.1 AGBW02011969 OWR45849.1 KQ459603 KPI92843.1 JXJN01022342 KQ971338 KYB27849.1 PYGN01001873 PSN32381.1 KB632278 ERL91250.1 GDHF01001397 JAI50917.1 GDHF01002047 JAI50267.1 CVRI01000054 CRL00722.1 AJVK01003260 GAMC01008527 JAB98028.1 JXUM01052978 KQ561757 KXJ77618.1 GAMC01008529 JAB98026.1 JRES01000986 KNC26429.1 DS231844 EDS35702.1 KK852424 KDR24197.1 CH963920 EDW77644.1 APGK01029087 APGK01029088 APGK01029089 APGK01029090 APGK01029091 APGK01029092 APGK01029093 APGK01029094 APGK01029095 APGK01029096 KB740724 ENN79361.1 CCAG010009664 AXCM01006694 CP012523 ALC39766.1 CH480820 EDW54214.1 APCN01002500 ADMH02001962 ETN60163.1 AXCN02000871 M80537 AE014134 CM002910 KMY88033.1 CH954177 EDV57707.2 KK855429 PTY22532.1 AAAB01008964 OUUW01000004 SPP79093.1 EAA12848.4 CH902620 EDV30764.2 CM000157 EDW87615.2 CH379059 EAL34175.3 KRT03716.1 ACPB03003610 CH940661 EDW71186.2 KQ435793 KOX74190.1 GL766908 EFZ14511.1 CH916368 EDW04285.1 KZ288203 PBC33405.1 PBC33406.1 KQ977041 KYN06149.1 KQ414617 KOC68171.1 GL434853 EFN74432.1 KQ981993 KYN30880.1 GL452770 EFN76851.1 GL732637 EFX69368.1 LBMM01000928 KMQ97200.1 QOIP01000007 RLU20895.1 GL888275 EGI63423.1 KQ976424 KYM88569.1 GDIP01188458 JAJ34944.1 ADTU01024312 ADTU01024313 ADTU01024314 ADTU01024315 ADTU01024316 ADTU01024317 ADTU01024318 ADTU01024319 ADTU01024320 ADTU01024321 GDIP01081671 JAM22044.1 GDIQ01001375 JAN93362.1 GDIP01039529 JAM64186.1 GDIP01225206 JAI98195.1 GDIP01188459 JAJ34943.1 GDIP01191058 JAJ32344.1 GDIP01231006 JAI92395.1 GDIP01236656 JAI86745.1 GDIP01120565 JAL83149.1 GDIP01227607 JAI95794.1 GDIP01250705 JAI72696.1 GDIP01227606 JAI95795.1 KQ981153 KYN09067.1 KQ982562 KYQ54945.1 DS235354 EEB15109.1 CH477394 EAT41892.1 ABJB010022594 ABJB010731548 ABJB010823282 ABJB010823901 ABJB010839736 DS668046 EEC03408.1 LJIJ01000407 ODM97827.1
JTDY01001533 KOB73626.1 AGBW02011969 OWR45849.1 KQ459603 KPI92843.1 JXJN01022342 KQ971338 KYB27849.1 PYGN01001873 PSN32381.1 KB632278 ERL91250.1 GDHF01001397 JAI50917.1 GDHF01002047 JAI50267.1 CVRI01000054 CRL00722.1 AJVK01003260 GAMC01008527 JAB98028.1 JXUM01052978 KQ561757 KXJ77618.1 GAMC01008529 JAB98026.1 JRES01000986 KNC26429.1 DS231844 EDS35702.1 KK852424 KDR24197.1 CH963920 EDW77644.1 APGK01029087 APGK01029088 APGK01029089 APGK01029090 APGK01029091 APGK01029092 APGK01029093 APGK01029094 APGK01029095 APGK01029096 KB740724 ENN79361.1 CCAG010009664 AXCM01006694 CP012523 ALC39766.1 CH480820 EDW54214.1 APCN01002500 ADMH02001962 ETN60163.1 AXCN02000871 M80537 AE014134 CM002910 KMY88033.1 CH954177 EDV57707.2 KK855429 PTY22532.1 AAAB01008964 OUUW01000004 SPP79093.1 EAA12848.4 CH902620 EDV30764.2 CM000157 EDW87615.2 CH379059 EAL34175.3 KRT03716.1 ACPB03003610 CH940661 EDW71186.2 KQ435793 KOX74190.1 GL766908 EFZ14511.1 CH916368 EDW04285.1 KZ288203 PBC33405.1 PBC33406.1 KQ977041 KYN06149.1 KQ414617 KOC68171.1 GL434853 EFN74432.1 KQ981993 KYN30880.1 GL452770 EFN76851.1 GL732637 EFX69368.1 LBMM01000928 KMQ97200.1 QOIP01000007 RLU20895.1 GL888275 EGI63423.1 KQ976424 KYM88569.1 GDIP01188458 JAJ34944.1 ADTU01024312 ADTU01024313 ADTU01024314 ADTU01024315 ADTU01024316 ADTU01024317 ADTU01024318 ADTU01024319 ADTU01024320 ADTU01024321 GDIP01081671 JAM22044.1 GDIQ01001375 JAN93362.1 GDIP01039529 JAM64186.1 GDIP01225206 JAI98195.1 GDIP01188459 JAJ34943.1 GDIP01191058 JAJ32344.1 GDIP01231006 JAI92395.1 GDIP01236656 JAI86745.1 GDIP01120565 JAL83149.1 GDIP01227607 JAI95794.1 GDIP01250705 JAI72696.1 GDIP01227606 JAI95795.1 KQ981153 KYN09067.1 KQ982562 KYQ54945.1 DS235354 EEB15109.1 CH477394 EAT41892.1 ABJB010022594 ABJB010731548 ABJB010823282 ABJB010823901 ABJB010839736 DS668046 EEC03408.1 LJIJ01000407 ODM97827.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000007151
UP000053268
+ More
UP000092460 UP000007266 UP000192223 UP000245037 UP000030742 UP000183832 UP000092462 UP000069940 UP000249989 UP000037069 UP000002320 UP000091820 UP000092443 UP000027135 UP000078200 UP000092445 UP000007798 UP000075881 UP000075885 UP000019118 UP000092444 UP000075883 UP000069272 UP000075902 UP000092553 UP000075920 UP000075903 UP000076407 UP000001292 UP000075840 UP000075900 UP000075901 UP000000673 UP000075886 UP000000803 UP000076408 UP000008711 UP000075880 UP000192221 UP000268350 UP000007062 UP000007801 UP000002282 UP000001819 UP000015103 UP000095301 UP000095300 UP000008792 UP000053105 UP000001070 UP000242457 UP000078542 UP000053825 UP000000311 UP000005203 UP000078541 UP000008237 UP000000305 UP000036403 UP000279307 UP000007755 UP000078540 UP000005205 UP000078492 UP000075809 UP000009046 UP000002358 UP000079169 UP000008820 UP000075882 UP000001555 UP000094527
UP000092460 UP000007266 UP000192223 UP000245037 UP000030742 UP000183832 UP000092462 UP000069940 UP000249989 UP000037069 UP000002320 UP000091820 UP000092443 UP000027135 UP000078200 UP000092445 UP000007798 UP000075881 UP000075885 UP000019118 UP000092444 UP000075883 UP000069272 UP000075902 UP000092553 UP000075920 UP000075903 UP000076407 UP000001292 UP000075840 UP000075900 UP000075901 UP000000673 UP000075886 UP000000803 UP000076408 UP000008711 UP000075880 UP000192221 UP000268350 UP000007062 UP000007801 UP000002282 UP000001819 UP000015103 UP000095301 UP000095300 UP000008792 UP000053105 UP000001070 UP000242457 UP000078542 UP000053825 UP000000311 UP000005203 UP000078541 UP000008237 UP000000305 UP000036403 UP000279307 UP000007755 UP000078540 UP000005205 UP000078492 UP000075809 UP000009046 UP000002358 UP000079169 UP000008820 UP000075882 UP000001555 UP000094527
Interpro
ProteinModelPortal
H9JM45
A0A2A4JGB0
A0A194R096
A0A0L7LEK0
A0A212EWJ7
A0A194PIG7
+ More
A0A1B0BXS9 A0A139WIU3 A0A1W4WS50 A0A2P8XK47 U4UMJ9 A0A0K8WIK8 A0A0K8WGR0 A0A1J1ILU1 A0A1B0D4J3 W8BM12 A0A182GBS5 W8BXI0 A0A0L0C4R4 B0W5P9 A0A1A9W0F1 A0A1A9XH88 A0A067RK74 A0A1A9UMV7 A0A1B0BXS8 A0A1A9ZJ37 B4N0N1 A0A182JZ92 A0A182PJ70 N6UFF5 A0A1B0FNU6 A0A182MKD8 A0A182F4L6 A0A182UAH5 A0A0M4E2E8 A0A182W7V5 A0A182VN95 A0A182WT87 B4I369 A0A182HV80 A0A182RBM1 A0A182SG56 W5J9S7 A0A182Q423 P33450 A0A0J9QVG6 A0A182YGU6 B3N3P8 A0A182IV90 A0A2R7WSJ6 A0A1W4UQG6 A0A1S4GXD7 A0A3B0JEL3 Q7Q3K5 B3MLP3 B4NYI2 Q29NP3 T1I617 A0A1I8MY28 A0A1I8P2A4 B4MDC5 A0A0M8ZZC1 E9IY59 B4JD41 A0A2A3ENR6 A0A2A3EQA1 A0A195D170 A0A0L7RBN5 E1ZWJ5 A0A088A7Z0 A0A195ERJ8 E2C5B8 E9HFS0 A0A0J7NXR0 A0A3L8DKB9 F4WQR0 A0A195BRJ0 A0A0P5CCQ8 A0A158NRY6 A0A0N8D072 A0A0P6J127 A0A0P5ZTU5 A0A0P4YMD5 A0A0P5B7A8 A0A0P5B1P8 A0A0P4YB10 A0A0P4XXI2 A0A0P5U2L5 A0A0P4ZCX0 A0A0N7ZFF4 A0A0P4YSZ5 A0A195D885 A0A151X3F5 E0VP03 K7ILV2 A0A3Q0JFL9 Q0IF47 A0A182L245 B7P9Y6 A0A1D2MXS1
A0A1B0BXS9 A0A139WIU3 A0A1W4WS50 A0A2P8XK47 U4UMJ9 A0A0K8WIK8 A0A0K8WGR0 A0A1J1ILU1 A0A1B0D4J3 W8BM12 A0A182GBS5 W8BXI0 A0A0L0C4R4 B0W5P9 A0A1A9W0F1 A0A1A9XH88 A0A067RK74 A0A1A9UMV7 A0A1B0BXS8 A0A1A9ZJ37 B4N0N1 A0A182JZ92 A0A182PJ70 N6UFF5 A0A1B0FNU6 A0A182MKD8 A0A182F4L6 A0A182UAH5 A0A0M4E2E8 A0A182W7V5 A0A182VN95 A0A182WT87 B4I369 A0A182HV80 A0A182RBM1 A0A182SG56 W5J9S7 A0A182Q423 P33450 A0A0J9QVG6 A0A182YGU6 B3N3P8 A0A182IV90 A0A2R7WSJ6 A0A1W4UQG6 A0A1S4GXD7 A0A3B0JEL3 Q7Q3K5 B3MLP3 B4NYI2 Q29NP3 T1I617 A0A1I8MY28 A0A1I8P2A4 B4MDC5 A0A0M8ZZC1 E9IY59 B4JD41 A0A2A3ENR6 A0A2A3EQA1 A0A195D170 A0A0L7RBN5 E1ZWJ5 A0A088A7Z0 A0A195ERJ8 E2C5B8 E9HFS0 A0A0J7NXR0 A0A3L8DKB9 F4WQR0 A0A195BRJ0 A0A0P5CCQ8 A0A158NRY6 A0A0N8D072 A0A0P6J127 A0A0P5ZTU5 A0A0P4YMD5 A0A0P5B7A8 A0A0P5B1P8 A0A0P4YB10 A0A0P4XXI2 A0A0P5U2L5 A0A0P4ZCX0 A0A0N7ZFF4 A0A0P4YSZ5 A0A195D885 A0A151X3F5 E0VP03 K7ILV2 A0A3Q0JFL9 Q0IF47 A0A182L245 B7P9Y6 A0A1D2MXS1
PDB
5T9T
E-value=4.98016e-29,
Score=318
Ontologies
PATHWAY
GO
GO:0007156
GO:0005886
GO:0005509
GO:0016021
GO:0098609
GO:0005911
GO:0045198
GO:0048105
GO:0045317
GO:0035209
GO:0016339
GO:0090251
GO:0035332
GO:0044331
GO:0018149
GO:0045296
GO:0007447
GO:0035159
GO:0008283
GO:0005887
GO:0010629
GO:0008285
GO:0043065
GO:0007446
GO:0016327
GO:0016336
GO:0032880
GO:0000904
GO:0090176
GO:0045571
GO:0001737
GO:0007157
GO:0045570
GO:0005739
GO:0001736
GO:0046621
GO:0016324
GO:0007164
GO:0045926
GO:0009888
GO:0060071
GO:0040008
GO:0042067
GO:0007476
GO:0035220
GO:0008353
GO:0004693
GO:0004022
GO:0007155
GO:0016020
GO:0006508
GO:0008237
GO:0043401
GO:0003707
GO:0051537
Topology
Subcellular location
Cell membrane
Apical cell membrane
Mitochondrion
Apical cell membrane
Mitochondrion
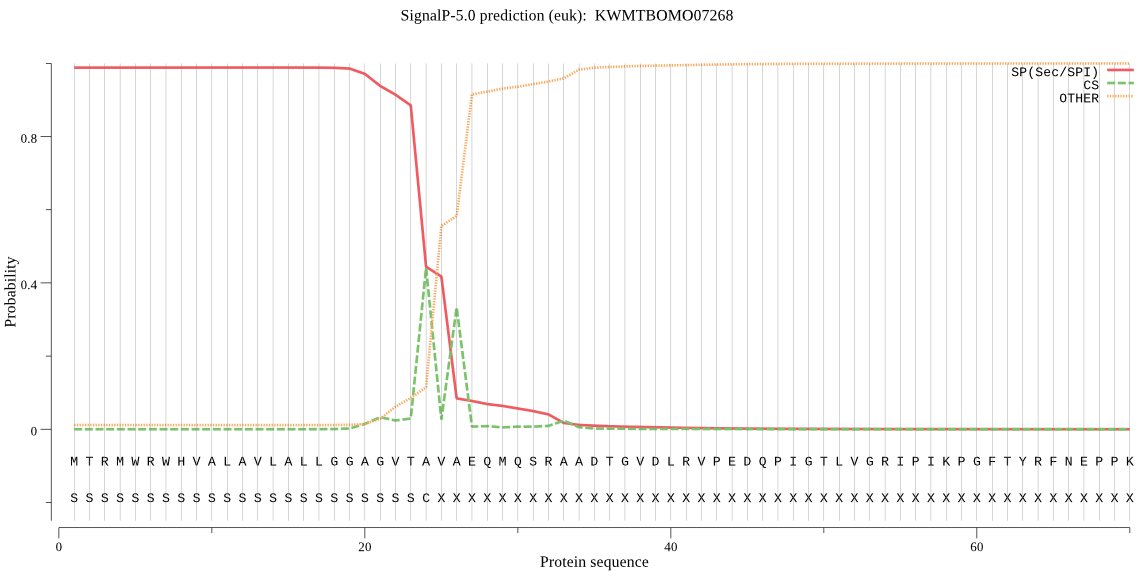
SignalP
Position: 1 - 24,
Likelihood: 0.988360
Length:
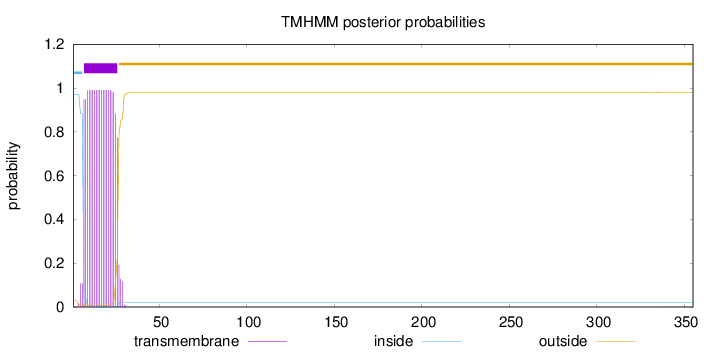
355
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.08842
Exp number, first 60 AAs:
20.07774
Total prob of N-in:
0.97145
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 26
outside
27 - 355
Population Genetic Test Statistics
Pi
8.310385
Theta
6.956201
Tajima's D
0.868534
CLR
0.221918
CSRT
0.644067796610169
Interpretation
Uncertain
