Gene
KWMTBOMO07266
Pre Gene Modal
BGIBMGA010592
Annotation
PREDICTED:_synaptic_vesicle_2-related_protein-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.944
Sequence
CDS
ATGCGTCGTGCCAAAGGATATCAGGATTTAGATGAGAGTGGAATTGCGGTTCGGTCTTTAGCGCCACCGATGCCGCCTCCACAAGAAATTGAAATGGCCTCAGTAACTGTAGTCCCTGACGACACATTCACAGTAACACAAGCTGTAAATGCTCTAGGTTTCGGTTGGTTCCAAGTGAAGCTATCATTATGCACAGGGCTCTGTTGGATGGCAGACTCAATGGAGATGACAATTCTAAGTATACTGTCACCAGCTTTACACTGCGAATGGAATATAAGCAAATACCAACAAGCTCTCACTACCACAGTTGTATTCATGGGTATGATGCTAAGCTCAACATTCTGGGGAAACATTAGTGATAGATATGGCAGAAAAACTGCTCTTAGTATGTGTGGCGTGCTCCTGTTTTACTATGGCCTGCTTAGCTCAATAGCACCAAATTTTCTTTGGTTACTCTTTTTAAGAGGCTTAGTTGGTTTTGCTATTGGATGTGTGCCCCAGTCTGTCACGTTGTATGCTGAATTTTTGCCAACCAAACAAAGAGCCAAATGTGTAGTATTGTTGGATTGTTTCTGGGCGTTAGGAGCATGTTTAGAAGTGGCACTCGCATTAGTCGTTATGCCGACTCTCGGCGTTCACTGGCTACTCGCTCTGTCTACAATACCCTTGCTTATGTTCGCACTCATATGCCCTTGGCTGCCAGAGTCGGCTCGATTTCATGTGGCTAGCGGGGAAGCTGATAAAGCACTTGCTACTCTTGAACAAATTGCACAAGACAACGGTCGACCCATGTTACTTGGGCGTTTGGTGTGCGATGATTCTGTGGGCATAGGTCAACGGGGAAGATTGAAGCACTTGCTCATACCGCATCTGAGGAATACTAGCCTTCTGCTGTGGTTTATATGGATGTCCTGTGCCTTCTGTTACTACGGCTTAGTCCTGATGGCGACAGAGTTGTTCGAATCGAATCCTGGTGGACTGGAGGAAGCGTGTGCCGCTGACTGCAGGCCGCTTCAGACGACAGATTACATGGACCTATTGTGGACTACACTAGCAGAGTTCCCGGGTATTTTCGCGACAATATTCATTATCGAGAAATTCGGTCGCAAACGAACAATGGCGACGCAGTTCATAATATTCGCTTTGTGCGTCTGCGTGTTGATTTATAACTCTGATCGAACATTCCTCACTTGCATTTTGTTCCTGGCTCGGGGTATTATAGCTGGACTCTTCCAAGCGGCCTATGTTTACACTCCAGAGGTGTACCCCACAGCGCTGAGAGCGACAGCGGTGGGCGCGTGCAGCGGGGTCGCTCGACTGGGAGCCATGATCACGCCCTACGTGGCACAGGTGTTACTAAAGAACTCGGTGGCTATAGCGACCGGGGTGTACGCATTGGCGGCGGTCTTAGCTGCCTTTGCCTGTCTCGCTCTACCCATCGAGACGAAAGGGAGAGAGATGCGAGACACAGTGACGACAGCTTCTTAA
Protein
MRRAKGYQDLDESGIAVRSLAPPMPPPQEIEMASVTVVPDDTFTVTQAVNALGFGWFQVKLSLCTGLCWMADSMEMTILSILSPALHCEWNISKYQQALTTTVVFMGMMLSSTFWGNISDRYGRKTALSMCGVLLFYYGLLSSIAPNFLWLLFLRGLVGFAIGCVPQSVTLYAEFLPTKQRAKCVVLLDCFWALGACLEVALALVVMPTLGVHWLLALSTIPLLMFALICPWLPESARFHVASGEADKALATLEQIAQDNGRPMLLGRLVCDDSVGIGQRGRLKHLLIPHLRNTSLLLWFIWMSCAFCYYGLVLMATELFESNPGGLEEACAADCRPLQTTDYMDLLWTTLAEFPGIFATIFIIEKFGRKRTMATQFIIFALCVCVLIYNSDRTFLTCILFLARGIIAGLFQAAYVYTPEVYPTALRATAVGACSGVARLGAMITPYVAQVLLKNSVAIATGVYALAAVLAAFACLALPIETKGREMRDTVTTAS
Summary
Uniprot
A0A194PHF3
A0A2A4JH16
H9JM40
A0A2H1VK12
A0A3S2LW25
A0A212EWK7
+ More
D6WME8 A0A1Y1KTK1 A0A194R0T7 A0A1W4WYI2 A0A067RC83 A0A2J7PKJ0 A0A1B6LHW8 A0A336M5F9 W5JKW5 A0A084VF85 E0VGL6 A0A182RB58 A0A182QAX0 A0A182XZG3 A0A182HEL0 A0A182FG00 A0A182WIU7 B0WVU0 A0A182NHG4 A0A182KRX4 A0A182P9W8 A0A182I3S5 A0A2M4BKD7 Q16J00 A0A2M4AG98 A0A2M4AGM2 A0A2M4AFE9 A0A182X6H2 A0A182UQJ5 A0A182U4H9 Q7QJU4 A0A2M4BJK8 A0A1B0CUK4 A0A182JWA3 A0A2H8TZP2 J9JQN7 K7ITS9 A0A1J1J878 A0A1B6F660 T1GU02 F4WBQ3 A0A195FSZ1 A0A154PB27 A0A158NRG9 A0A0L7R142 E2BC75 V9ICG1 A0A195DIE4 A0A195C2V6 A0A151XHQ0 E2AXE4 A0A195BE54 A0A026VT93 E9H0K8 A0A0P4W790 A0A310SYV4 A0A0P4XJC1 A0A0P6DTQ1 A0A0P4Y2S2 A0A0A1WGK5 A0A0P6B7D6 A0A0K8V205 A0A0P6HGN0 A0A2A3EH41 V9IER5 B4MQT2 T1JBX0 A0A1S3I3N1 A0A1S3I3K1 F6XK47 B4GCH3 B4QBG8 A0A210PRM8 Q290B2 A0A2C9KJL3 A0A2Y9ICP1 A0A3Q7QER9 A0A1S3EZG9 A0A3Q7RJG0 B4I8Z2 Q8SYM5 M3XWG3 A0A383ZYD4 U6CVY6 A0A0P6GNR7 G1MDL0 M3X573 A0A096MNK0 A0A341B6G6 A0A0A9ZAP7 A0A2M4AMX6 A0A340WFC1 A0A2U4CKZ7 A0A2Y9SMD7
D6WME8 A0A1Y1KTK1 A0A194R0T7 A0A1W4WYI2 A0A067RC83 A0A2J7PKJ0 A0A1B6LHW8 A0A336M5F9 W5JKW5 A0A084VF85 E0VGL6 A0A182RB58 A0A182QAX0 A0A182XZG3 A0A182HEL0 A0A182FG00 A0A182WIU7 B0WVU0 A0A182NHG4 A0A182KRX4 A0A182P9W8 A0A182I3S5 A0A2M4BKD7 Q16J00 A0A2M4AG98 A0A2M4AGM2 A0A2M4AFE9 A0A182X6H2 A0A182UQJ5 A0A182U4H9 Q7QJU4 A0A2M4BJK8 A0A1B0CUK4 A0A182JWA3 A0A2H8TZP2 J9JQN7 K7ITS9 A0A1J1J878 A0A1B6F660 T1GU02 F4WBQ3 A0A195FSZ1 A0A154PB27 A0A158NRG9 A0A0L7R142 E2BC75 V9ICG1 A0A195DIE4 A0A195C2V6 A0A151XHQ0 E2AXE4 A0A195BE54 A0A026VT93 E9H0K8 A0A0P4W790 A0A310SYV4 A0A0P4XJC1 A0A0P6DTQ1 A0A0P4Y2S2 A0A0A1WGK5 A0A0P6B7D6 A0A0K8V205 A0A0P6HGN0 A0A2A3EH41 V9IER5 B4MQT2 T1JBX0 A0A1S3I3N1 A0A1S3I3K1 F6XK47 B4GCH3 B4QBG8 A0A210PRM8 Q290B2 A0A2C9KJL3 A0A2Y9ICP1 A0A3Q7QER9 A0A1S3EZG9 A0A3Q7RJG0 B4I8Z2 Q8SYM5 M3XWG3 A0A383ZYD4 U6CVY6 A0A0P6GNR7 G1MDL0 M3X573 A0A096MNK0 A0A341B6G6 A0A0A9ZAP7 A0A2M4AMX6 A0A340WFC1 A0A2U4CKZ7 A0A2Y9SMD7
Pubmed
26354079
19121390
22118469
18362917
19820115
28004739
+ More
24845553 20920257 23761445 24438588 20566863 25244985 26483478 20966253 17510324 12364791 20075255 21719571 21347285 20798317 24508170 30249741 21292972 25830018 17994087 16341006 22936249 28812685 15632085 23185243 15562597 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20010809 17975172 25401762
24845553 20920257 23761445 24438588 20566863 25244985 26483478 20966253 17510324 12364791 20075255 21719571 21347285 20798317 24508170 30249741 21292972 25830018 17994087 16341006 22936249 28812685 15632085 23185243 15562597 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20010809 17975172 25401762
EMBL
KQ459603
KPI92841.1
NWSH01001485
PCG71109.1
BABH01039932
BABH01039933
+ More
BABH01039934 BABH01039935 ODYU01002725 SOQ40584.1 RSAL01000165 RVE45284.1 AGBW02011969 OWR45851.1 KQ971343 EFA03313.2 GEZM01075870 JAV63908.1 KQ460883 KPJ11124.1 KK852732 KDR17464.1 NEVH01024540 PNF16829.1 GEBQ01016665 JAT23312.1 UFQT01000369 SSX23607.1 ADMH02001234 ETN63555.1 ATLV01012365 ATLV01012366 KE524785 KFB36629.1 DS235149 EEB12522.1 AXCN02000591 JXUM01035613 JXUM01035614 JXUM01035615 KQ561067 KXJ79813.1 DS232132 EDS35772.1 APCN01000220 GGFJ01004117 MBW53258.1 CH478040 EAT34254.1 GGFK01006436 MBW39757.1 GGFK01006612 MBW39933.1 GGFK01006198 MBW39519.1 AAAB01008807 EAA04631.3 GGFJ01004118 MBW53259.1 AJWK01029294 AJWK01029295 GFXV01007575 MBW19380.1 ABLF02025352 CVRI01000075 CRL08603.1 GECZ01024105 JAS45664.1 CAQQ02037815 CAQQ02037816 GL888066 EGI68331.1 KQ981280 KYN43417.1 KQ434850 KZC08584.1 ADTU01023989 KQ414668 KOC64568.1 GL447256 EFN86702.1 JR039728 AEY58818.1 KQ980824 KYN12602.1 KQ978317 KYM95194.1 KQ982123 KYQ59877.1 GL443548 EFN61924.1 KQ976511 KYM82477.1 KK108116 QOIP01000005 EZA46890.1 RLU22617.1 GL732581 EFX74666.1 GDRN01077457 JAI62739.1 KQ759767 OAD62967.1 GDIP01240486 JAI82915.1 GDIQ01072510 LRGB01001005 JAN22227.1 KZS14128.1 GDIP01233437 JAI89964.1 GBXI01016662 JAC97629.1 GDIP01020932 JAM82783.1 GDHF01019516 JAI32798.1 GDIQ01019087 JAN75650.1 KZ288254 PBC30784.1 JR039729 AEY58819.1 CH963849 EDW74471.1 JH432025 AAEX03014752 CH479181 EDW32450.1 CM000362 CM002911 EDX08486.1 KMY96214.1 NEDP02005541 OWF39141.1 CM000071 EAL25450.1 KRT02234.1 CH480824 EDW57067.1 AE013599 AY071447 AAF47135.2 AAL49069.1 AEYP01095843 AEYP01095844 AEYP01095845 AEYP01095846 AEYP01095847 HAAF01002633 CCP74459.1 GDIQ01041219 JAN53518.1 ACTA01107918 ACTA01115918 AANG04002472 AHZZ02002942 AHZZ02002943 AHZZ02002944 GBHO01038454 GBHO01003176 JAG05150.1 JAG40428.1 GGFK01008637 MBW41958.1
BABH01039934 BABH01039935 ODYU01002725 SOQ40584.1 RSAL01000165 RVE45284.1 AGBW02011969 OWR45851.1 KQ971343 EFA03313.2 GEZM01075870 JAV63908.1 KQ460883 KPJ11124.1 KK852732 KDR17464.1 NEVH01024540 PNF16829.1 GEBQ01016665 JAT23312.1 UFQT01000369 SSX23607.1 ADMH02001234 ETN63555.1 ATLV01012365 ATLV01012366 KE524785 KFB36629.1 DS235149 EEB12522.1 AXCN02000591 JXUM01035613 JXUM01035614 JXUM01035615 KQ561067 KXJ79813.1 DS232132 EDS35772.1 APCN01000220 GGFJ01004117 MBW53258.1 CH478040 EAT34254.1 GGFK01006436 MBW39757.1 GGFK01006612 MBW39933.1 GGFK01006198 MBW39519.1 AAAB01008807 EAA04631.3 GGFJ01004118 MBW53259.1 AJWK01029294 AJWK01029295 GFXV01007575 MBW19380.1 ABLF02025352 CVRI01000075 CRL08603.1 GECZ01024105 JAS45664.1 CAQQ02037815 CAQQ02037816 GL888066 EGI68331.1 KQ981280 KYN43417.1 KQ434850 KZC08584.1 ADTU01023989 KQ414668 KOC64568.1 GL447256 EFN86702.1 JR039728 AEY58818.1 KQ980824 KYN12602.1 KQ978317 KYM95194.1 KQ982123 KYQ59877.1 GL443548 EFN61924.1 KQ976511 KYM82477.1 KK108116 QOIP01000005 EZA46890.1 RLU22617.1 GL732581 EFX74666.1 GDRN01077457 JAI62739.1 KQ759767 OAD62967.1 GDIP01240486 JAI82915.1 GDIQ01072510 LRGB01001005 JAN22227.1 KZS14128.1 GDIP01233437 JAI89964.1 GBXI01016662 JAC97629.1 GDIP01020932 JAM82783.1 GDHF01019516 JAI32798.1 GDIQ01019087 JAN75650.1 KZ288254 PBC30784.1 JR039729 AEY58819.1 CH963849 EDW74471.1 JH432025 AAEX03014752 CH479181 EDW32450.1 CM000362 CM002911 EDX08486.1 KMY96214.1 NEDP02005541 OWF39141.1 CM000071 EAL25450.1 KRT02234.1 CH480824 EDW57067.1 AE013599 AY071447 AAF47135.2 AAL49069.1 AEYP01095843 AEYP01095844 AEYP01095845 AEYP01095846 AEYP01095847 HAAF01002633 CCP74459.1 GDIQ01041219 JAN53518.1 ACTA01107918 ACTA01115918 AANG04002472 AHZZ02002942 AHZZ02002943 AHZZ02002944 GBHO01038454 GBHO01003176 JAG05150.1 JAG40428.1 GGFK01008637 MBW41958.1
Proteomes
UP000053268
UP000218220
UP000005204
UP000283053
UP000007151
UP000007266
+ More
UP000053240 UP000192223 UP000027135 UP000235965 UP000000673 UP000030765 UP000009046 UP000075900 UP000075886 UP000076408 UP000069940 UP000249989 UP000069272 UP000075920 UP000002320 UP000075884 UP000075882 UP000075885 UP000075840 UP000008820 UP000076407 UP000075903 UP000075902 UP000007062 UP000092461 UP000075881 UP000007819 UP000002358 UP000183832 UP000015102 UP000007755 UP000078541 UP000076502 UP000005205 UP000053825 UP000008237 UP000078492 UP000078542 UP000075809 UP000000311 UP000078540 UP000053097 UP000279307 UP000000305 UP000076858 UP000242457 UP000007798 UP000085678 UP000002254 UP000008744 UP000000304 UP000242188 UP000001819 UP000076420 UP000248481 UP000286641 UP000081671 UP000286640 UP000001292 UP000000803 UP000000715 UP000261681 UP000008912 UP000011712 UP000028761 UP000252040 UP000265300 UP000245320
UP000053240 UP000192223 UP000027135 UP000235965 UP000000673 UP000030765 UP000009046 UP000075900 UP000075886 UP000076408 UP000069940 UP000249989 UP000069272 UP000075920 UP000002320 UP000075884 UP000075882 UP000075885 UP000075840 UP000008820 UP000076407 UP000075903 UP000075902 UP000007062 UP000092461 UP000075881 UP000007819 UP000002358 UP000183832 UP000015102 UP000007755 UP000078541 UP000076502 UP000005205 UP000053825 UP000008237 UP000078492 UP000078542 UP000075809 UP000000311 UP000078540 UP000053097 UP000279307 UP000000305 UP000076858 UP000242457 UP000007798 UP000085678 UP000002254 UP000008744 UP000000304 UP000242188 UP000001819 UP000076420 UP000248481 UP000286641 UP000081671 UP000286640 UP000001292 UP000000803 UP000000715 UP000261681 UP000008912 UP000011712 UP000028761 UP000252040 UP000265300 UP000245320
Interpro
CDD
ProteinModelPortal
A0A194PHF3
A0A2A4JH16
H9JM40
A0A2H1VK12
A0A3S2LW25
A0A212EWK7
+ More
D6WME8 A0A1Y1KTK1 A0A194R0T7 A0A1W4WYI2 A0A067RC83 A0A2J7PKJ0 A0A1B6LHW8 A0A336M5F9 W5JKW5 A0A084VF85 E0VGL6 A0A182RB58 A0A182QAX0 A0A182XZG3 A0A182HEL0 A0A182FG00 A0A182WIU7 B0WVU0 A0A182NHG4 A0A182KRX4 A0A182P9W8 A0A182I3S5 A0A2M4BKD7 Q16J00 A0A2M4AG98 A0A2M4AGM2 A0A2M4AFE9 A0A182X6H2 A0A182UQJ5 A0A182U4H9 Q7QJU4 A0A2M4BJK8 A0A1B0CUK4 A0A182JWA3 A0A2H8TZP2 J9JQN7 K7ITS9 A0A1J1J878 A0A1B6F660 T1GU02 F4WBQ3 A0A195FSZ1 A0A154PB27 A0A158NRG9 A0A0L7R142 E2BC75 V9ICG1 A0A195DIE4 A0A195C2V6 A0A151XHQ0 E2AXE4 A0A195BE54 A0A026VT93 E9H0K8 A0A0P4W790 A0A310SYV4 A0A0P4XJC1 A0A0P6DTQ1 A0A0P4Y2S2 A0A0A1WGK5 A0A0P6B7D6 A0A0K8V205 A0A0P6HGN0 A0A2A3EH41 V9IER5 B4MQT2 T1JBX0 A0A1S3I3N1 A0A1S3I3K1 F6XK47 B4GCH3 B4QBG8 A0A210PRM8 Q290B2 A0A2C9KJL3 A0A2Y9ICP1 A0A3Q7QER9 A0A1S3EZG9 A0A3Q7RJG0 B4I8Z2 Q8SYM5 M3XWG3 A0A383ZYD4 U6CVY6 A0A0P6GNR7 G1MDL0 M3X573 A0A096MNK0 A0A341B6G6 A0A0A9ZAP7 A0A2M4AMX6 A0A340WFC1 A0A2U4CKZ7 A0A2Y9SMD7
D6WME8 A0A1Y1KTK1 A0A194R0T7 A0A1W4WYI2 A0A067RC83 A0A2J7PKJ0 A0A1B6LHW8 A0A336M5F9 W5JKW5 A0A084VF85 E0VGL6 A0A182RB58 A0A182QAX0 A0A182XZG3 A0A182HEL0 A0A182FG00 A0A182WIU7 B0WVU0 A0A182NHG4 A0A182KRX4 A0A182P9W8 A0A182I3S5 A0A2M4BKD7 Q16J00 A0A2M4AG98 A0A2M4AGM2 A0A2M4AFE9 A0A182X6H2 A0A182UQJ5 A0A182U4H9 Q7QJU4 A0A2M4BJK8 A0A1B0CUK4 A0A182JWA3 A0A2H8TZP2 J9JQN7 K7ITS9 A0A1J1J878 A0A1B6F660 T1GU02 F4WBQ3 A0A195FSZ1 A0A154PB27 A0A158NRG9 A0A0L7R142 E2BC75 V9ICG1 A0A195DIE4 A0A195C2V6 A0A151XHQ0 E2AXE4 A0A195BE54 A0A026VT93 E9H0K8 A0A0P4W790 A0A310SYV4 A0A0P4XJC1 A0A0P6DTQ1 A0A0P4Y2S2 A0A0A1WGK5 A0A0P6B7D6 A0A0K8V205 A0A0P6HGN0 A0A2A3EH41 V9IER5 B4MQT2 T1JBX0 A0A1S3I3N1 A0A1S3I3K1 F6XK47 B4GCH3 B4QBG8 A0A210PRM8 Q290B2 A0A2C9KJL3 A0A2Y9ICP1 A0A3Q7QER9 A0A1S3EZG9 A0A3Q7RJG0 B4I8Z2 Q8SYM5 M3XWG3 A0A383ZYD4 U6CVY6 A0A0P6GNR7 G1MDL0 M3X573 A0A096MNK0 A0A341B6G6 A0A0A9ZAP7 A0A2M4AMX6 A0A340WFC1 A0A2U4CKZ7 A0A2Y9SMD7
PDB
4J05
E-value=9.01542e-11,
Score=162
Ontologies
GO
Topology
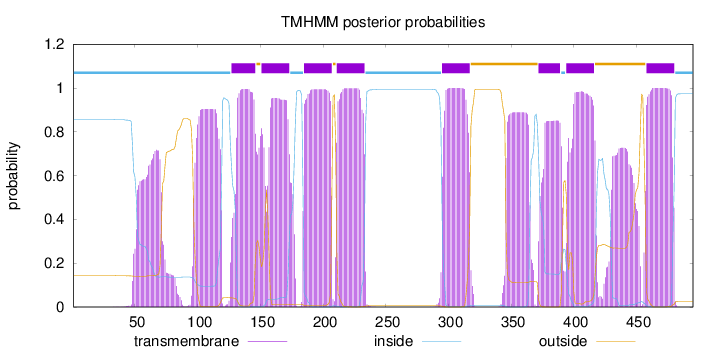
Length:
495
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
237.51897
Exp number, first 60 AAs:
6.20433
Total prob of N-in:
0.85683
inside
1 - 126
TMhelix
127 - 146
outside
147 - 150
TMhelix
151 - 173
inside
174 - 184
TMhelix
185 - 207
outside
208 - 210
TMhelix
211 - 233
inside
234 - 294
TMhelix
295 - 317
outside
318 - 371
TMhelix
372 - 389
inside
390 - 393
TMhelix
394 - 416
outside
417 - 457
TMhelix
458 - 480
inside
481 - 495
Population Genetic Test Statistics
Pi
25.185749
Theta
21.173469
Tajima's D
-1.428878
CLR
0.86466
CSRT
0.0660466976651167
Interpretation
Uncertain
