Pre Gene Modal
BGIBMGA010356
Annotation
PREDICTED:_synaptotagmin_I_isoform_X3_[Bombyx_mori]
Full name
Synaptotagmin 1
Alternative Name
p65
Location in the cell
Cytoplasmic Reliability : 1.522 Nuclear Reliability : 1.928
Sequence
CDS
ATGTCACCGGCTCTGGGCACGGTGCTGCGATGGCGCCGCGAAGCCGGCCCTCCAGAGGCCGTCGTCGAGGATGTTAAAGAGACAAGTCCACGCCCAGAAAATACTACCGAATCCGTGATGACTCCATTAGCATCAACTACAATGGCTCCAACTCAAGCGTCTATAGGTGAACAAGCTGCGAACCTAGCGCGGGGAATGAATTTGGAGACGTGGCAGCTGGTCGCCATTCTTGTGGGTGTCGGTGTGGTGATTCTGGGGATATGTTTCTGCTGCATCAGGAGATGTTTCCGAAAGCGACGCTCCAAGGATGGCAAGAAAGGGATGAAGGGTGTCGATCTAAAATCCGTCCAGCTGTTGGGTTCAGCATACAAAGAAAAGGTTCAGCCTGATATGGAGGAGCTCACGGAAAACGCTGAGGAACCCGACGATGGTGATTCCAAAAAAGAGGAACAAAAGCTCGGAAAACTTCAGTACAAGCTGGAATATGACTTCAACAGTAACAGTTTGTCAGTGACCGTAATCCAGGCTGAGGACTTACCGGCCTTGGACATGGGCGGAACATCTGACCCTTACGTGAAAGTATATCTTCTTCCGGATAAGAAGAAGAAATTCGAAACTAAAGTTCACCGAAAAACATTGAGCCCTGTTTTCAATGAGACGTTCGTGTTCAAGAACGTGCCATACGCTGACGCAATGAACAAGACCCTTGTATTCGCCATTTTCGATTTTGATCGCTTCTCCAAACACGATCAAATCGGTGAGGTGAAGGTACCATTATGTCAAGTGGATTTGGCACAGACAATCGAGGAATGGCGTGAGCTTCAGAGTGTTGAAGGTGAAGGAGGTCAGCTGGGAGACATTTGTTTCTCATTGCGTTACGTACCTACGGCTGGAAAACTAACTGTTGTCATACTGGAAGCCAAGAATCTAAAGAAAATGGACGTCGGTGGTCTCTCAGATCCGTACGTAAAGATTGCACTTATGCAAAACGGCAAACGTCTGAAGAAGAAGAAAACAAGCATCAAGAAATGCACGCTGAATCCCTATTACAACGAGTCATTTACTTTTGAAGTACCATTCGAACAAATACAGAAAGTGAATCTAGTAATCACCGTGGTGGATTATGATCGTATTGGCACATCGGAGCCTATTGGCAAAGTTGTGCTTGGTTACAACGCGTCGGGAACTGAACTACGCCACTGGTCCGACATGCTGGCCAGCCCGCGACGCCCCATCGCTCAGTGGCACACCCTTAAGGATCCCGATGATGGAGAAAAGAAGGACTAA
Protein
MSPALGTVLRWRREAGPPEAVVEDVKETSPRPENTTESVMTPLASTTMAPTQASIGEQAANLARGMNLETWQLVAILVGVGVVILGICFCCIRRCFRKRRSKDGKKGMKGVDLKSVQLLGSAYKEKVQPDMEELTENAEEPDDGDSKKEEQKLGKLQYKLEYDFNSNSLSVTVIQAEDLPALDMGGTSDPYVKVYLLPDKKKKFETKVHRKTLSPVFNETFVFKNVPYADAMNKTLVFAIFDFDRFSKHDQIGEVKVPLCQVDLAQTIEEWRELQSVEGEGGQLGDICFSLRYVPTAGKLTVVILEAKNLKKMDVGGLSDPYVKIALMQNGKRLKKKKTSIKKCTLNPYYNESFTFEVPFEQIQKVNLVITVVDYDRIGTSEPIGKVVLGYNASGTELRHWSDMLASPRRPIAQWHTLKDPDDGEKKD
Summary
Description
May have a regulatory role in the membrane interactions during trafficking of synaptic vesicles at the active zone of the synapse. It binds acidic phospholipids with a specificity that requires the presence of both an acidic head group and a diacyl backbone.
Cofactor
Ca(2+)
Subunit
Homodimer or homotrimer (Potential). Identified in a complex with Syn and nwk (PubMed:29568072). Interacts with StnA and StnB via its second C2 domain. This interaction may mediate its retrieval from the plasma membrane, thereby facilitating the internalization of multiple synaptic vesicles from the plasma membrane.
Similarity
Belongs to the synaptotagmin family.
Keywords
Alternative splicing
Calcium
Cell junction
Complete proteome
Cytoplasmic vesicle
Membrane
Metal-binding
Reference proteome
Repeat
RNA editing
Synapse
Transmembrane
Transmembrane helix
Feature
chain Synaptotagmin 1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
C0K3M8
A0A2A4IZR1
Q9BMF3
A0A2W1BZI3
A0A3S2KZA2
A0A076FE67
+ More
A0A194PHE8 A0A212FG40 A0A1Q3FRC3 U5ERC4 A0A1Q3FRF6 A0A1Q3FRK6 D2A0P6 A0A1W4X2Z9 A0A088AT06 C0K3M5 B0WCF7 A0A0L7KSK1 E2AE99 C0K3M7 A0A2A3EG78 C0K3M9 V9I7G8 A0A182V3U5 A0A151WS27 A0A151IIL1 A0A151IIM1 A0A0J7L5A4 A0A151ISN2 F4X438 A0A151JXM3 A0A158NE18 A0A151I0A6 A0A026WUQ5 A0A2R7VQR1 E2BJ45 A0A232EWP1 A0A1Q3FQU7 A0A182PJ84 Q7PN22 A0A182MVQ6 A0A182NEB5 A0A084VP08 A0A0C9QQ67 E9IA62 C0K3M6 A0A154NW04 A0A1Y1NB03 F5HSC2 A0A0T6AXT7 A0A023F5S9 A0A1Y1NAG9 K7J3M7 A0A1B6KUS8 A0A1B6CUQ6 A0A1B6IIF8 A0A1B6FF82 A0A336M1C6 A0A1Q3FRN3 A0A2H8TDH8 A0A182YGT0 J9JXJ7 A0A1Q3FRH2 A0A0K8S8S9 A0A1Q3FRP4 U4UD35 A0A0A9YV49 A0A1Q3FRB5 A0A0L7QUV5 A0A1Q3FRU3 A0A1W4V6A6 A0A182WT68 A0A0J9QUN2 A0A182HV63 P21521-4 A0A0R1DIT0 A0A182L222 A0A0Q5W952 A0A1Q3FQY5 A0A1S4GWR3 A0A182U4K5 A0A3B0JNB7 A0A0Q9X836 A0A182RBK5 A0A182K1L2 A0A1I8MD79 A0A0P8XYP0 A0A182F4K1 A0A0Q9WRQ8 A0A0Q9WL46 A0A1W4V5W6 X2J979 B4NX83 B3NA62 P21521 B4I2Q5 B4Q8K7 B4KLJ2 A0A0K8U299 A0A0P6ALJ6
A0A194PHE8 A0A212FG40 A0A1Q3FRC3 U5ERC4 A0A1Q3FRF6 A0A1Q3FRK6 D2A0P6 A0A1W4X2Z9 A0A088AT06 C0K3M5 B0WCF7 A0A0L7KSK1 E2AE99 C0K3M7 A0A2A3EG78 C0K3M9 V9I7G8 A0A182V3U5 A0A151WS27 A0A151IIL1 A0A151IIM1 A0A0J7L5A4 A0A151ISN2 F4X438 A0A151JXM3 A0A158NE18 A0A151I0A6 A0A026WUQ5 A0A2R7VQR1 E2BJ45 A0A232EWP1 A0A1Q3FQU7 A0A182PJ84 Q7PN22 A0A182MVQ6 A0A182NEB5 A0A084VP08 A0A0C9QQ67 E9IA62 C0K3M6 A0A154NW04 A0A1Y1NB03 F5HSC2 A0A0T6AXT7 A0A023F5S9 A0A1Y1NAG9 K7J3M7 A0A1B6KUS8 A0A1B6CUQ6 A0A1B6IIF8 A0A1B6FF82 A0A336M1C6 A0A1Q3FRN3 A0A2H8TDH8 A0A182YGT0 J9JXJ7 A0A1Q3FRH2 A0A0K8S8S9 A0A1Q3FRP4 U4UD35 A0A0A9YV49 A0A1Q3FRB5 A0A0L7QUV5 A0A1Q3FRU3 A0A1W4V6A6 A0A182WT68 A0A0J9QUN2 A0A182HV63 P21521-4 A0A0R1DIT0 A0A182L222 A0A0Q5W952 A0A1Q3FQY5 A0A1S4GWR3 A0A182U4K5 A0A3B0JNB7 A0A0Q9X836 A0A182RBK5 A0A182K1L2 A0A1I8MD79 A0A0P8XYP0 A0A182F4K1 A0A0Q9WRQ8 A0A0Q9WL46 A0A1W4V5W6 X2J979 B4NX83 B3NA62 P21521 B4I2Q5 B4Q8K7 B4KLJ2 A0A0K8U299 A0A0P6ALJ6
Pubmed
19121390
28756777
25683246
26354079
22118469
18362917
+ More
19820115 19221008 26227816 20798317 21719571 21347285 24508170 30249741 28648823 12364791 14747013 17210077 24438588 21282665 28004739 25474469 20075255 25244985 23537049 25401762 26823975 22936249 1840599 10731132 12537572 12537569 11069931 12907802 29568072 17994087 17550304 20966253 25315136 12537568 12537573 12537574 16110336 17569856 17569867
19820115 19221008 26227816 20798317 21719571 21347285 24508170 30249741 28648823 12364791 14747013 17210077 24438588 21282665 28004739 25474469 20075255 25244985 23537049 25401762 26823975 22936249 1840599 10731132 12537572 12537569 11069931 12907802 29568072 17994087 17550304 20966253 25315136 12537568 12537573 12537574 16110336 17569856 17569867
EMBL
BABH01005848
FJ550351
ACN21657.1
NWSH01004301
PCG65241.1
AF331039
+ More
AAK01129.1 KZ149911 PZC78126.1 RSAL01000381 RVE42056.1 KJ863735 AII15699.1 KQ459603 KPI92836.1 AGBW02008720 OWR52699.1 GFDL01004906 JAV30139.1 GANO01002866 JAB57005.1 GFDL01004908 JAV30137.1 GFDL01004933 JAV30112.1 KQ971338 EFA02813.1 FJ550348 ACN21654.1 DS231887 EDS43481.1 JTDY01006398 KOB66046.1 GL438827 EFN68332.1 FJ550350 ACN21656.1 KZ288254 PBC30737.1 FJ550352 ACN21658.1 JR036247 JR036249 AEY57030.1 AEY57031.1 KQ982796 KYQ50597.1 KQ977477 KYN02491.1 KYN02490.1 LBMM01000694 KMQ97728.1 KQ981064 KYN09846.1 GL888625 EGI58762.1 KQ981607 KYN39414.1 ADTU01013109 ADTU01013110 KQ976687 KYM78142.1 KK107098 QOIP01000001 EZA59693.1 RLU26947.1 KK854019 PTY09498.1 GL448558 EFN84195.1 NNAY01001843 OXU22745.1 GFDL01005106 JAV29939.1 AAAB01008964 EAA12894.5 AXCM01005688 ATLV01014966 KE524999 KFB39702.1 GBYB01002772 GBYB01002774 JAG72539.1 JAG72541.1 GL761987 EFZ22520.1 FJ550349 ACN21655.1 KQ434771 KZC03827.1 GEZM01007793 JAV94919.1 AB630173 BAK23253.1 LJIG01022570 KRT79874.1 GBBI01002180 JAC16532.1 GEZM01007791 JAV94922.1 GEBQ01024782 JAT15195.1 GEDC01020078 JAS17220.1 GECU01033136 GECU01020994 JAS74570.1 JAS86712.1 GECZ01020897 JAS48872.1 UFQT01000409 SSX24032.1 GFDL01004899 JAV30146.1 GFXV01000339 MBW12144.1 ABLF02032715 ABLF02032718 ABLF02032721 ABLF02032722 GFDL01004900 JAV30145.1 GBRD01016138 JAG49688.1 GFDL01004893 JAV30152.1 KB632266 ERL90967.1 GBHO01006697 GDHC01010853 JAG36907.1 JAQ07776.1 GFDL01004977 JAV30068.1 KQ414731 KOC62400.1 GFDL01004758 JAV30287.1 CM002910 KMY87741.1 APCN01002494 M55048 AE014134 BT004498 AY119455 CM000157 KRJ97157.1 CH954177 KQS70012.1 GFDL01005025 JAV30020.1 OUUW01000006 SPP81842.1 CH933807 KRG03465.1 CH902652 KPU74624.1 CH963913 KRF98649.1 CH940649 KRF81563.1 AHN54082.1 EDW87440.1 EDV57525.1 CH480820 EDW54050.1 CM000361 EDX03524.1 KMY87736.1 EDW12873.1 GDHF01031445 JAI20869.1 GDIP01027800 JAM75915.1
AAK01129.1 KZ149911 PZC78126.1 RSAL01000381 RVE42056.1 KJ863735 AII15699.1 KQ459603 KPI92836.1 AGBW02008720 OWR52699.1 GFDL01004906 JAV30139.1 GANO01002866 JAB57005.1 GFDL01004908 JAV30137.1 GFDL01004933 JAV30112.1 KQ971338 EFA02813.1 FJ550348 ACN21654.1 DS231887 EDS43481.1 JTDY01006398 KOB66046.1 GL438827 EFN68332.1 FJ550350 ACN21656.1 KZ288254 PBC30737.1 FJ550352 ACN21658.1 JR036247 JR036249 AEY57030.1 AEY57031.1 KQ982796 KYQ50597.1 KQ977477 KYN02491.1 KYN02490.1 LBMM01000694 KMQ97728.1 KQ981064 KYN09846.1 GL888625 EGI58762.1 KQ981607 KYN39414.1 ADTU01013109 ADTU01013110 KQ976687 KYM78142.1 KK107098 QOIP01000001 EZA59693.1 RLU26947.1 KK854019 PTY09498.1 GL448558 EFN84195.1 NNAY01001843 OXU22745.1 GFDL01005106 JAV29939.1 AAAB01008964 EAA12894.5 AXCM01005688 ATLV01014966 KE524999 KFB39702.1 GBYB01002772 GBYB01002774 JAG72539.1 JAG72541.1 GL761987 EFZ22520.1 FJ550349 ACN21655.1 KQ434771 KZC03827.1 GEZM01007793 JAV94919.1 AB630173 BAK23253.1 LJIG01022570 KRT79874.1 GBBI01002180 JAC16532.1 GEZM01007791 JAV94922.1 GEBQ01024782 JAT15195.1 GEDC01020078 JAS17220.1 GECU01033136 GECU01020994 JAS74570.1 JAS86712.1 GECZ01020897 JAS48872.1 UFQT01000409 SSX24032.1 GFDL01004899 JAV30146.1 GFXV01000339 MBW12144.1 ABLF02032715 ABLF02032718 ABLF02032721 ABLF02032722 GFDL01004900 JAV30145.1 GBRD01016138 JAG49688.1 GFDL01004893 JAV30152.1 KB632266 ERL90967.1 GBHO01006697 GDHC01010853 JAG36907.1 JAQ07776.1 GFDL01004977 JAV30068.1 KQ414731 KOC62400.1 GFDL01004758 JAV30287.1 CM002910 KMY87741.1 APCN01002494 M55048 AE014134 BT004498 AY119455 CM000157 KRJ97157.1 CH954177 KQS70012.1 GFDL01005025 JAV30020.1 OUUW01000006 SPP81842.1 CH933807 KRG03465.1 CH902652 KPU74624.1 CH963913 KRF98649.1 CH940649 KRF81563.1 AHN54082.1 EDW87440.1 EDV57525.1 CH480820 EDW54050.1 CM000361 EDX03524.1 KMY87736.1 EDW12873.1 GDHF01031445 JAI20869.1 GDIP01027800 JAM75915.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000007266
+ More
UP000192223 UP000005203 UP000002320 UP000037510 UP000000311 UP000242457 UP000075903 UP000075809 UP000078542 UP000036403 UP000078492 UP000007755 UP000078541 UP000005205 UP000078540 UP000053097 UP000279307 UP000008237 UP000215335 UP000075885 UP000007062 UP000075883 UP000075884 UP000030765 UP000076502 UP000002358 UP000076408 UP000007819 UP000030742 UP000053825 UP000192221 UP000076407 UP000075840 UP000000803 UP000002282 UP000075882 UP000008711 UP000075902 UP000268350 UP000009192 UP000075900 UP000075881 UP000095301 UP000007801 UP000069272 UP000007798 UP000008792 UP000001292 UP000000304
UP000192223 UP000005203 UP000002320 UP000037510 UP000000311 UP000242457 UP000075903 UP000075809 UP000078542 UP000036403 UP000078492 UP000007755 UP000078541 UP000005205 UP000078540 UP000053097 UP000279307 UP000008237 UP000215335 UP000075885 UP000007062 UP000075883 UP000075884 UP000030765 UP000076502 UP000002358 UP000076408 UP000007819 UP000030742 UP000053825 UP000192221 UP000076407 UP000075840 UP000000803 UP000002282 UP000075882 UP000008711 UP000075902 UP000268350 UP000009192 UP000075900 UP000075881 UP000095301 UP000007801 UP000069272 UP000007798 UP000008792 UP000001292 UP000000304
Interpro
Gene 3D
ProteinModelPortal
C0K3M8
A0A2A4IZR1
Q9BMF3
A0A2W1BZI3
A0A3S2KZA2
A0A076FE67
+ More
A0A194PHE8 A0A212FG40 A0A1Q3FRC3 U5ERC4 A0A1Q3FRF6 A0A1Q3FRK6 D2A0P6 A0A1W4X2Z9 A0A088AT06 C0K3M5 B0WCF7 A0A0L7KSK1 E2AE99 C0K3M7 A0A2A3EG78 C0K3M9 V9I7G8 A0A182V3U5 A0A151WS27 A0A151IIL1 A0A151IIM1 A0A0J7L5A4 A0A151ISN2 F4X438 A0A151JXM3 A0A158NE18 A0A151I0A6 A0A026WUQ5 A0A2R7VQR1 E2BJ45 A0A232EWP1 A0A1Q3FQU7 A0A182PJ84 Q7PN22 A0A182MVQ6 A0A182NEB5 A0A084VP08 A0A0C9QQ67 E9IA62 C0K3M6 A0A154NW04 A0A1Y1NB03 F5HSC2 A0A0T6AXT7 A0A023F5S9 A0A1Y1NAG9 K7J3M7 A0A1B6KUS8 A0A1B6CUQ6 A0A1B6IIF8 A0A1B6FF82 A0A336M1C6 A0A1Q3FRN3 A0A2H8TDH8 A0A182YGT0 J9JXJ7 A0A1Q3FRH2 A0A0K8S8S9 A0A1Q3FRP4 U4UD35 A0A0A9YV49 A0A1Q3FRB5 A0A0L7QUV5 A0A1Q3FRU3 A0A1W4V6A6 A0A182WT68 A0A0J9QUN2 A0A182HV63 P21521-4 A0A0R1DIT0 A0A182L222 A0A0Q5W952 A0A1Q3FQY5 A0A1S4GWR3 A0A182U4K5 A0A3B0JNB7 A0A0Q9X836 A0A182RBK5 A0A182K1L2 A0A1I8MD79 A0A0P8XYP0 A0A182F4K1 A0A0Q9WRQ8 A0A0Q9WL46 A0A1W4V5W6 X2J979 B4NX83 B3NA62 P21521 B4I2Q5 B4Q8K7 B4KLJ2 A0A0K8U299 A0A0P6ALJ6
A0A194PHE8 A0A212FG40 A0A1Q3FRC3 U5ERC4 A0A1Q3FRF6 A0A1Q3FRK6 D2A0P6 A0A1W4X2Z9 A0A088AT06 C0K3M5 B0WCF7 A0A0L7KSK1 E2AE99 C0K3M7 A0A2A3EG78 C0K3M9 V9I7G8 A0A182V3U5 A0A151WS27 A0A151IIL1 A0A151IIM1 A0A0J7L5A4 A0A151ISN2 F4X438 A0A151JXM3 A0A158NE18 A0A151I0A6 A0A026WUQ5 A0A2R7VQR1 E2BJ45 A0A232EWP1 A0A1Q3FQU7 A0A182PJ84 Q7PN22 A0A182MVQ6 A0A182NEB5 A0A084VP08 A0A0C9QQ67 E9IA62 C0K3M6 A0A154NW04 A0A1Y1NB03 F5HSC2 A0A0T6AXT7 A0A023F5S9 A0A1Y1NAG9 K7J3M7 A0A1B6KUS8 A0A1B6CUQ6 A0A1B6IIF8 A0A1B6FF82 A0A336M1C6 A0A1Q3FRN3 A0A2H8TDH8 A0A182YGT0 J9JXJ7 A0A1Q3FRH2 A0A0K8S8S9 A0A1Q3FRP4 U4UD35 A0A0A9YV49 A0A1Q3FRB5 A0A0L7QUV5 A0A1Q3FRU3 A0A1W4V6A6 A0A182WT68 A0A0J9QUN2 A0A182HV63 P21521-4 A0A0R1DIT0 A0A182L222 A0A0Q5W952 A0A1Q3FQY5 A0A1S4GWR3 A0A182U4K5 A0A3B0JNB7 A0A0Q9X836 A0A182RBK5 A0A182K1L2 A0A1I8MD79 A0A0P8XYP0 A0A182F4K1 A0A0Q9WRQ8 A0A0Q9WL46 A0A1W4V5W6 X2J979 B4NX83 B3NA62 P21521 B4I2Q5 B4Q8K7 B4KLJ2 A0A0K8U299 A0A0P6ALJ6
PDB
5KJ8
E-value=6.17862e-111,
Score=1025
Ontologies
GO
GO:0008021
GO:0005544
GO:0016021
GO:0007269
GO:0048791
GO:0030424
GO:0030276
GO:0016079
GO:0070382
GO:0005509
GO:0001786
GO:0005886
GO:0014059
GO:0031045
GO:0019905
GO:0030672
GO:0017158
GO:0016192
GO:0017156
GO:0071277
GO:0000149
GO:0048488
GO:0016020
GO:0031594
GO:0030054
GO:0043195
GO:0008345
GO:0042803
GO:0030285
GO:0060024
GO:0007317
GO:0150007
GO:0050803
GO:0007268
GO:0031340
GO:1900073
GO:0005515
GO:0051056
GO:0030170
GO:0006520
GO:0006508
GO:0008237
GO:0043401
GO:0003707
GO:0051537
PANTHER
Topology
Subcellular location
Cytoplasmic vesicle
Secretory vesicle
Synaptic vesicle membrane
Cell junction
Synapse
Secretory vesicle
Synaptic vesicle membrane
Cell junction
Synapse
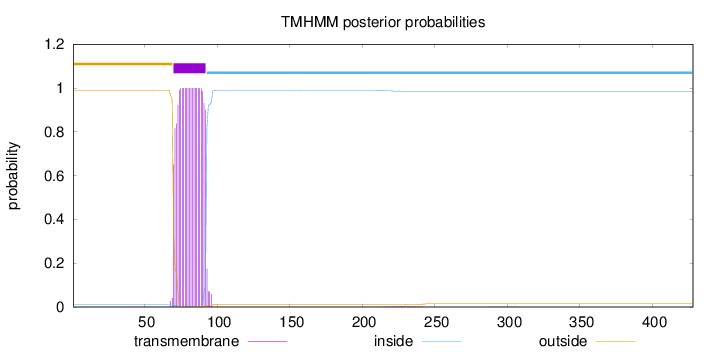
Length:
428
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.5639499999999
Exp number, first 60 AAs:
0.00641
Total prob of N-in:
0.01054
outside
1 - 69
TMhelix
70 - 92
inside
93 - 428
Population Genetic Test Statistics
Pi
4.204475
Theta
22.743111
Tajima's D
-0.573027
CLR
0
CSRT
0.220838958052097
Interpretation
Uncertain
