Gene
KWMTBOMO07260
Annotation
LYR_motif-containing_protein_5_[Operophtera_brumata]
Full name
Electron transfer flavoprotein regulatory factor 1
Alternative Name
LYR motif-containing protein 5
Growth hormone-inducible soluble protein
Growth hormone-inducible soluble protein
Location in the cell
Cytoplasmic Reliability : 1.487 Mitochondrial Reliability : 1.782
Sequence
CDS
ATGGCCCAGCCACATAGAAAAGCGGTTTTGAATCTTTATAAAACGCTCCTCTATCTGGGTCGAGATTGGCCTCAAGGTTTCGACTTGTTTAGAAAAAGGCTTCATAAAGTATTTAGCAAGAATAGTGAAGAAACTGATCCACAAAAAATACAAATAATGGTAAAACATGGTGAGTTTGTTGTTAAAGAAATTGAAGCACTATATAAGTTGAAGAAGTACAGAGCAATGAAGAGACGATATTATGACGATAACTAA
Protein
MAQPHRKAVLNLYKTLLYLGRDWPQGFDLFRKRLHKVFSKNSEETDPQKIQIMVKHGEFVVKEIEALYKLKKYRAMKRRYYDDN
Summary
Description
Acts as a regulator of the electron transfer flavoprotein by promoting the removal of flavin from the ETF holoenzyme (composed of ETFA and ETFB).
Subunit
homotetramer. Interacts with NDUFAB1. Interacts with ETFA. Interacts with ETFB.
Similarity
Belongs to the complex I LYR family.
Keywords
Complete proteome
Mitochondrion
Reference proteome
Polymorphism
Feature
chain Electron transfer flavoprotein regulatory factor 1
Uniprot
A0A0L7KRY9
A0A212FG44
A0A2H1WDQ6
A0A194R1L1
A0A194PNY2
S4PFK0
+ More
U4UF67 A0A067QRH5 A0A0V0G9C0 A0A232EZZ2 A0A3S2NK56 K7J3L9 C3KHN2 A0A1S2ZE35 A0A2A3EG86 A0A1B6MJK7 A0A1S3FVG0 A0A069DNJ7 B5FXA0 A0A224Y5G5 A0A2U9B1T0 A0A0P7XST2 H0ZPS2 A0A3B3S0T3 J9JZU2 H2SF26 F7FN02 A0A1W4WS85 A0A1W4X371 A0A218UH55 F4WS65 A0A1A7YNN0 A0A091F8Z8 A0A3P8W8V8 A0A3Q3DRF9 A0A1W4X305 A0A151NIW5 A0A1U7RX22 A0A1B0CZ95 A0A2S2QTY1 E2AE50 H3CN90 A0A3P8RWN0 A0A151IC61 A0A3Q1B920 E0VZF5 A0A3B4Z5R4 A0A1S3D5Z6 H3C2T5 Q4SSR5 U3JMM0 A0A151ISN4 A0A2G9S515 A0A1A8RAJ5 A0A1A8M8H3 A0A1A8KMH8 A0A1A8D6M3 A0A1A8AUA6 A0A3Q3EET3 A0A2J7PVI2 A0A139WJD6 A0A0M9A382 Q91V16 A0A3B3XJS2 A0A091HSK8 A0A3Q1FZ63 A0A226CW98 A0A087Y7B6 D4AD58 A0A3Q0RJ79 A0A151JW03 A0A026WBY2 G3H3D0 A0A3P8X4S1 A0A182VQG8 A0A182MMQ4 E2BQ91 A0A3Q2E1R8 A0A1A6G2G4 A0A3L7H6D1 A0A1U7Q8Q2 A0A3Q3W6K5 A0A3P9Q7G2 A0A3L8SGZ2 G3T1H8 A0A2A6CJ09 A0A2J7PTC1 A0A182IRL7 A0A0S7MAW8 A0A3N0YCJ4 A0A3B4A0U9 A0A2J7NKH5 A0A087XBI5 A0A3S2PN67 A0A091PA59 A0A0Q3U127 A0A3B3UAQ7 H3BD02 A0A3Q3RKC3 A0A3B5RDD3
U4UF67 A0A067QRH5 A0A0V0G9C0 A0A232EZZ2 A0A3S2NK56 K7J3L9 C3KHN2 A0A1S2ZE35 A0A2A3EG86 A0A1B6MJK7 A0A1S3FVG0 A0A069DNJ7 B5FXA0 A0A224Y5G5 A0A2U9B1T0 A0A0P7XST2 H0ZPS2 A0A3B3S0T3 J9JZU2 H2SF26 F7FN02 A0A1W4WS85 A0A1W4X371 A0A218UH55 F4WS65 A0A1A7YNN0 A0A091F8Z8 A0A3P8W8V8 A0A3Q3DRF9 A0A1W4X305 A0A151NIW5 A0A1U7RX22 A0A1B0CZ95 A0A2S2QTY1 E2AE50 H3CN90 A0A3P8RWN0 A0A151IC61 A0A3Q1B920 E0VZF5 A0A3B4Z5R4 A0A1S3D5Z6 H3C2T5 Q4SSR5 U3JMM0 A0A151ISN4 A0A2G9S515 A0A1A8RAJ5 A0A1A8M8H3 A0A1A8KMH8 A0A1A8D6M3 A0A1A8AUA6 A0A3Q3EET3 A0A2J7PVI2 A0A139WJD6 A0A0M9A382 Q91V16 A0A3B3XJS2 A0A091HSK8 A0A3Q1FZ63 A0A226CW98 A0A087Y7B6 D4AD58 A0A3Q0RJ79 A0A151JW03 A0A026WBY2 G3H3D0 A0A3P8X4S1 A0A182VQG8 A0A182MMQ4 E2BQ91 A0A3Q2E1R8 A0A1A6G2G4 A0A3L7H6D1 A0A1U7Q8Q2 A0A3Q3W6K5 A0A3P9Q7G2 A0A3L8SGZ2 G3T1H8 A0A2A6CJ09 A0A2J7PTC1 A0A182IRL7 A0A0S7MAW8 A0A3N0YCJ4 A0A3B4A0U9 A0A2J7NKH5 A0A087XBI5 A0A3S2PN67 A0A091PA59 A0A0Q3U127 A0A3B3UAQ7 H3BD02 A0A3Q3RKC3 A0A3B5RDD3
Pubmed
26227816
22118469
26354079
23622113
23537049
24845553
+ More
28648823 20075255 26334808 17018643 20360741 29240929 21551351 18464734 21719571 24487278 22293439 20798317 15496914 20566863 18362917 19820115 16141072 19468303 15489334 15064703 21183079 15057822 15632090 24508170 30249741 21804562 29704459 30282656 18806794 25463417 9215903 23542700
28648823 20075255 26334808 17018643 20360741 29240929 21551351 18464734 21719571 24487278 22293439 20798317 15496914 20566863 18362917 19820115 16141072 19468303 15489334 15064703 21183079 15057822 15632090 24508170 30249741 21804562 29704459 30282656 18806794 25463417 9215903 23542700
EMBL
JTDY01006398
KOB66048.1
AGBW02008720
OWR52700.1
ODYU01007912
SOQ51106.1
+ More
KQ460883 KPJ11130.1 KQ459603 KPI92835.1 GAIX01006550 JAA86010.1 KB632266 ERL90968.1 KK853046 KDR12104.1 GECL01001764 JAP04360.1 NNAY01001475 OXU23837.1 RSAL01000381 RVE42055.1 BT082447 ACQ58154.1 KZ288254 PBC30747.1 GEBQ01019656 GEBQ01003830 JAT20321.1 JAT36147.1 GBGD01003653 JAC85236.1 DQ213190 GFTR01000697 JAW15729.1 CP026244 AWO97897.1 AWO97899.1 JARO02000060 KPP80090.1 ABQF01036320 ABLF02017840 ABLF02017843 ABLF02017844 ABLF02028087 ABLF02042871 MUZQ01000306 OWK53064.1 GL888300 EGI62959.1 HADW01006120 HADX01009522 SBP31754.1 KK719749 KFO65711.1 AKHW03002956 KYO36737.1 AJVK01002156 GGMS01011981 MBY81184.1 GL438827 EFN68283.1 KQ978067 KYM97670.1 DS235852 EEB18761.1 CAAE01014347 CAF96317.1 AGTO01001588 KQ981064 KYN09854.1 KV925902 PIO35232.1 HAEH01015257 HAEI01013769 SBS02274.1 HAEF01011958 HAEG01010950 SBR53117.1 HAED01008139 HAEE01013418 SBR33468.1 HADZ01020643 HAEA01001420 SBQ29900.1 HADY01019335 HAEJ01003791 SBP57820.1 NEVH01020958 PNF20344.1 KQ971338 KYB28063.1 KQ435750 KOX76345.1 AF412298 AF412299 AF412300 AY753325 AK015530 CU207316 BC021522 AY490388 KL217730 KFO98122.1 LNIX01000061 OXA37253.1 AYCK01011940 AABR07062484 CH473964 EDM01440.1 KQ981672 KYN38425.1 KK107292 QOIP01000001 EZA53483.1 RLU27133.1 JH000125 EGV97862.1 AXCM01016227 GL449698 EFN82145.1 LZPO01107928 OBS59990.1 RAZU01000269 RLQ61506.1 QUSF01000021 RLW01869.1 ABKE03000026 PDM78068.1 NEVH01021599 PNF19576.1 GBYX01087757 JAO98796.1 RJVU01047120 ROL43947.1 NEVH01053300 PNE09474.1 AYCK01012164 CM012459 RVE56311.1 KK847034 KFP88425.1 LMAW01000281 KQL59670.1 AFYH01038991
KQ460883 KPJ11130.1 KQ459603 KPI92835.1 GAIX01006550 JAA86010.1 KB632266 ERL90968.1 KK853046 KDR12104.1 GECL01001764 JAP04360.1 NNAY01001475 OXU23837.1 RSAL01000381 RVE42055.1 BT082447 ACQ58154.1 KZ288254 PBC30747.1 GEBQ01019656 GEBQ01003830 JAT20321.1 JAT36147.1 GBGD01003653 JAC85236.1 DQ213190 GFTR01000697 JAW15729.1 CP026244 AWO97897.1 AWO97899.1 JARO02000060 KPP80090.1 ABQF01036320 ABLF02017840 ABLF02017843 ABLF02017844 ABLF02028087 ABLF02042871 MUZQ01000306 OWK53064.1 GL888300 EGI62959.1 HADW01006120 HADX01009522 SBP31754.1 KK719749 KFO65711.1 AKHW03002956 KYO36737.1 AJVK01002156 GGMS01011981 MBY81184.1 GL438827 EFN68283.1 KQ978067 KYM97670.1 DS235852 EEB18761.1 CAAE01014347 CAF96317.1 AGTO01001588 KQ981064 KYN09854.1 KV925902 PIO35232.1 HAEH01015257 HAEI01013769 SBS02274.1 HAEF01011958 HAEG01010950 SBR53117.1 HAED01008139 HAEE01013418 SBR33468.1 HADZ01020643 HAEA01001420 SBQ29900.1 HADY01019335 HAEJ01003791 SBP57820.1 NEVH01020958 PNF20344.1 KQ971338 KYB28063.1 KQ435750 KOX76345.1 AF412298 AF412299 AF412300 AY753325 AK015530 CU207316 BC021522 AY490388 KL217730 KFO98122.1 LNIX01000061 OXA37253.1 AYCK01011940 AABR07062484 CH473964 EDM01440.1 KQ981672 KYN38425.1 KK107292 QOIP01000001 EZA53483.1 RLU27133.1 JH000125 EGV97862.1 AXCM01016227 GL449698 EFN82145.1 LZPO01107928 OBS59990.1 RAZU01000269 RLQ61506.1 QUSF01000021 RLW01869.1 ABKE03000026 PDM78068.1 NEVH01021599 PNF19576.1 GBYX01087757 JAO98796.1 RJVU01047120 ROL43947.1 NEVH01053300 PNE09474.1 AYCK01012164 CM012459 RVE56311.1 KK847034 KFP88425.1 LMAW01000281 KQL59670.1 AFYH01038991
Proteomes
UP000037510
UP000007151
UP000053240
UP000053268
UP000030742
UP000027135
+ More
UP000215335 UP000283053 UP000002358 UP000079721 UP000242457 UP000081671 UP000007754 UP000246464 UP000034805 UP000192224 UP000261540 UP000007819 UP000005226 UP000002279 UP000192223 UP000197619 UP000007755 UP000052976 UP000265120 UP000264820 UP000050525 UP000189705 UP000092462 UP000000311 UP000007303 UP000265080 UP000078542 UP000257160 UP000009046 UP000261400 UP000079169 UP000016665 UP000078492 UP000261660 UP000235965 UP000007266 UP000053105 UP000000589 UP000261480 UP000054308 UP000257200 UP000198287 UP000028760 UP000002494 UP000261340 UP000078541 UP000053097 UP000279307 UP000001075 UP000075920 UP000075883 UP000008237 UP000265020 UP000092124 UP000273346 UP000189706 UP000261620 UP000242638 UP000276834 UP000007646 UP000232936 UP000075880 UP000261520 UP000051836 UP000261500 UP000008672 UP000261640 UP000002852
UP000215335 UP000283053 UP000002358 UP000079721 UP000242457 UP000081671 UP000007754 UP000246464 UP000034805 UP000192224 UP000261540 UP000007819 UP000005226 UP000002279 UP000192223 UP000197619 UP000007755 UP000052976 UP000265120 UP000264820 UP000050525 UP000189705 UP000092462 UP000000311 UP000007303 UP000265080 UP000078542 UP000257160 UP000009046 UP000261400 UP000079169 UP000016665 UP000078492 UP000261660 UP000235965 UP000007266 UP000053105 UP000000589 UP000261480 UP000054308 UP000257200 UP000198287 UP000028760 UP000002494 UP000261340 UP000078541 UP000053097 UP000279307 UP000001075 UP000075920 UP000075883 UP000008237 UP000265020 UP000092124 UP000273346 UP000189706 UP000261620 UP000242638 UP000276834 UP000007646 UP000232936 UP000075880 UP000261520 UP000051836 UP000261500 UP000008672 UP000261640 UP000002852
Pfam
PF05347 Complex1_LYR
Interpro
IPR008011
Complex1_LYR
ProteinModelPortal
A0A0L7KRY9
A0A212FG44
A0A2H1WDQ6
A0A194R1L1
A0A194PNY2
S4PFK0
+ More
U4UF67 A0A067QRH5 A0A0V0G9C0 A0A232EZZ2 A0A3S2NK56 K7J3L9 C3KHN2 A0A1S2ZE35 A0A2A3EG86 A0A1B6MJK7 A0A1S3FVG0 A0A069DNJ7 B5FXA0 A0A224Y5G5 A0A2U9B1T0 A0A0P7XST2 H0ZPS2 A0A3B3S0T3 J9JZU2 H2SF26 F7FN02 A0A1W4WS85 A0A1W4X371 A0A218UH55 F4WS65 A0A1A7YNN0 A0A091F8Z8 A0A3P8W8V8 A0A3Q3DRF9 A0A1W4X305 A0A151NIW5 A0A1U7RX22 A0A1B0CZ95 A0A2S2QTY1 E2AE50 H3CN90 A0A3P8RWN0 A0A151IC61 A0A3Q1B920 E0VZF5 A0A3B4Z5R4 A0A1S3D5Z6 H3C2T5 Q4SSR5 U3JMM0 A0A151ISN4 A0A2G9S515 A0A1A8RAJ5 A0A1A8M8H3 A0A1A8KMH8 A0A1A8D6M3 A0A1A8AUA6 A0A3Q3EET3 A0A2J7PVI2 A0A139WJD6 A0A0M9A382 Q91V16 A0A3B3XJS2 A0A091HSK8 A0A3Q1FZ63 A0A226CW98 A0A087Y7B6 D4AD58 A0A3Q0RJ79 A0A151JW03 A0A026WBY2 G3H3D0 A0A3P8X4S1 A0A182VQG8 A0A182MMQ4 E2BQ91 A0A3Q2E1R8 A0A1A6G2G4 A0A3L7H6D1 A0A1U7Q8Q2 A0A3Q3W6K5 A0A3P9Q7G2 A0A3L8SGZ2 G3T1H8 A0A2A6CJ09 A0A2J7PTC1 A0A182IRL7 A0A0S7MAW8 A0A3N0YCJ4 A0A3B4A0U9 A0A2J7NKH5 A0A087XBI5 A0A3S2PN67 A0A091PA59 A0A0Q3U127 A0A3B3UAQ7 H3BD02 A0A3Q3RKC3 A0A3B5RDD3
U4UF67 A0A067QRH5 A0A0V0G9C0 A0A232EZZ2 A0A3S2NK56 K7J3L9 C3KHN2 A0A1S2ZE35 A0A2A3EG86 A0A1B6MJK7 A0A1S3FVG0 A0A069DNJ7 B5FXA0 A0A224Y5G5 A0A2U9B1T0 A0A0P7XST2 H0ZPS2 A0A3B3S0T3 J9JZU2 H2SF26 F7FN02 A0A1W4WS85 A0A1W4X371 A0A218UH55 F4WS65 A0A1A7YNN0 A0A091F8Z8 A0A3P8W8V8 A0A3Q3DRF9 A0A1W4X305 A0A151NIW5 A0A1U7RX22 A0A1B0CZ95 A0A2S2QTY1 E2AE50 H3CN90 A0A3P8RWN0 A0A151IC61 A0A3Q1B920 E0VZF5 A0A3B4Z5R4 A0A1S3D5Z6 H3C2T5 Q4SSR5 U3JMM0 A0A151ISN4 A0A2G9S515 A0A1A8RAJ5 A0A1A8M8H3 A0A1A8KMH8 A0A1A8D6M3 A0A1A8AUA6 A0A3Q3EET3 A0A2J7PVI2 A0A139WJD6 A0A0M9A382 Q91V16 A0A3B3XJS2 A0A091HSK8 A0A3Q1FZ63 A0A226CW98 A0A087Y7B6 D4AD58 A0A3Q0RJ79 A0A151JW03 A0A026WBY2 G3H3D0 A0A3P8X4S1 A0A182VQG8 A0A182MMQ4 E2BQ91 A0A3Q2E1R8 A0A1A6G2G4 A0A3L7H6D1 A0A1U7Q8Q2 A0A3Q3W6K5 A0A3P9Q7G2 A0A3L8SGZ2 G3T1H8 A0A2A6CJ09 A0A2J7PTC1 A0A182IRL7 A0A0S7MAW8 A0A3N0YCJ4 A0A3B4A0U9 A0A2J7NKH5 A0A087XBI5 A0A3S2PN67 A0A091PA59 A0A0Q3U127 A0A3B3UAQ7 H3BD02 A0A3Q3RKC3 A0A3B5RDD3
Ontologies
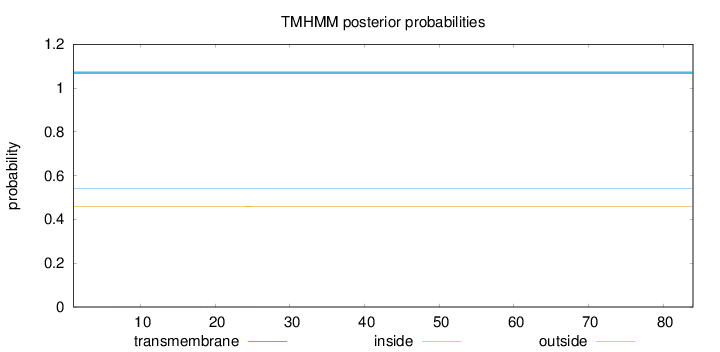
Topology
Subcellular location
Mitochondrion
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00524999999999999
Exp number, first 60 AAs:
0.00516999999999999
Total prob of N-in:
0.54026
inside
1 - 84
Population Genetic Test Statistics
Pi
0.717196
Theta
4.111594
Tajima's D
-2.215991
CLR
26.05047
CSRT
0.00294985250737463
Interpretation
Uncertain
