Gene
KWMTBOMO07255
Pre Gene Modal
BGIBMGA010358
Annotation
PREDICTED:_signal-induced_proliferation-associated_1-like_protein_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.266
Sequence
CDS
ATGTATGCTCCAGACCAGAGAAGAAAGGAAACGAGAGATGTTATATATCGCAGCAATTCGAGCCTTGACTTGATATACACCGAGCATTCCACCGGAAACGGCTTAAGACGTGAATATGGAAGTCACGGATCTATCGACGTGATAGGTGGAGCAAACGACAGATTTCCAGCAATGGTTCATAATTACAATGGAATATCAACGCCTGAAAAGCATCCAGGCTTAACCAAAGCGAATGAGCGCTTTACTGAAGTGAACAGCGATCATGCAGACGGACCCACGCCCCGTTCAAGTGCCAGTCCCAAAACTAGACTGAAACTAAACAAACTGTGGGGTGCAGCCTCTGGAACCCGACCACAACTCGATGAAGTATCAGTCTCAAGTGTTACAGATGTTTCTAAAAGGAAGCAATTTGCTCACTATGACTGTCAATCATTAATGACTAATTTGAATTATGCTGCTAAAGTCCGTGGTGAACTACTTAGTCGTAGACGCAACACAACTACTGGTGCATCTGCTGCATCTGCAACCAGAGGACTTCCATCAGGTGAAGATGCTTTAGGAGACACAGGGGATGGTAGATCAAATGAGCTACTTGAGAGTTGTCCGTACTTCAGGAACGAGCTAGGAGGGGAAGAAGAACGATGTGTGTCATTATGTTGGCGTACTGCACGTGGAGCTATCATAGGACAGGCGGGTGGTTGGCATCGGCCACGGACGTCGTACGGTATTTCAATGCTGGAGTTCCCACCGGGTCATTCACATTGGCGAAACGGCTGCCCTTTCGTTAGGTCAATGGCACCACTTCCAATCGAAAGTCAAGATCTAGGTGCTTTATACTATCGAAACTATTTCTACAACCAACCGCACCAGAACTGGTTCGGAATGGATGACATCCTCGGACCAGTAGCAGTTTCCATAAAGAAAGAACGTGTTGAATTACGACGTGGCAACACAGACGCATCGACGCCCCTATCCCAACTCGTATGGCAATACCGCCTACTCATACGGACTTCCGAATTGCTCACCTTACGCGGATCCATACTCGAAGATGCTTTAATAAATATTAAACAAACAAACAGCGCTTCTTATAACACAAAAGAAGTTTTGGAATATGTTGCGCCGGAGTTACAACTTAACTGCCTAAGACTGGGGATCTCAAACAGAGGTGCAGAAGAGCAACTATTACGATTAGACGAACAATGCGTAACACGACATTATAAAGTAGGAGTTATGTACTGCAAAAGTGGCCAATCAACTGAAGAAGAAATGTATAACAACCAGGAAGCTGGCCCAGCATTTGTTGAATTCCTTCAAATGTTGGGGCAAACCGTTAGATTGAAAGATTTCGACAAGTACAAGGCTGGCTTAGACAATAAAACTGATTCGACCGGGCTGTATTCTGTGTTCAGTACTTATCAAAGCTGCGACATTATGTTTCACGTGTCAACGATGCTTCCCTACACACCGAATAACAGACAGCAACTGTTAAGAAAACGACATATCGGAAACGATATCGTAACTATAGTATTTCAGGAGCCCGGCGCCGCCCCGTTCACTCCGCGCAATATTCGTTCACAGTTTCAACATGTCTTTATAGTTGTCCGAGCAATAGATCCATGTACGGACCACACTCACTACAGTATAGCTGTTAGTCGTGCTAAAGAAGTGCCTTTATTTGGCCCCCCGATAAAAGACGGTGCTGTTTATCCCAAAGGAGAAGCCTTTACCGATTTGCTTTTAAGTAAGGTAATAAATGGCGAAAATGCTGCAATACAATCACCAAAGTTTAGTACAATGGCAACACGTACTCGCCAAGAGTATCTAAAAGACCTTGCGAAGAATTACATCACAGCTACCACAGTTGAAACCGGTCAAAAGTTTTCATTATTCTCATCATTGCTAATGAAAAGTGGAAGTAATTCCGCGAACAGCACGATGGTAGCTCGCGTGGATAACTGTACTAACGACGTTCTGGTGGGCGGTGCGCTTTGCTTTCATGTTCATGTGCAAGATGAAGGTGCTGGTGGAACATTAGTAGAATGTATAATGGGAATTTCGGCTGATACTATAGTGTTTGTTGAAGATTTAAATCGACAAATTATCCTAGTTTTGCCAACTGCTTCGCTGCTTGGTTTCCATGTCCAACCAGATGGCTTAGTGACGGCAGTCGAAGGCGGAGGGTGTGCAGATAGAGCTGGCATACGTCGCGGCGCCCGGCTGCTGGAGGTGTGCCGCGCCGCTGTTGTAGCGCTCAACCATGATCAACTCGTAGATCTGCTCAAAACATCTGACCCGGTCACTGTGCGAGTAACGTTCGCGTCGGGTCTGAAGTGTCAGTGCGAGACCGGCGGCGAGGAGTGGGCGGCGCAGGCGCGGGCCGACGCGCAGCAGCCGGCGCGCGGCCGCCGGGCCTACGAGCGCGGCCACTCGCCGCCCAGATCCGACAGTTCCGGATACGGCACAGGAGGATCGGGTCGCAGCGGTCGACGGTCGCCGGACGGTGTGCGGCGCCGCTCGCATGACGACTCGTCGCCGCGCCACTCTCGGCAGAAACGCTCCTCCGCTAATACCAGCTCGCTTACGGAGGAACTGATGCGGCTGATGGACGCGGAGCTGGGGCCGGGGCGCCTGGAGAGCAGCGCGGCGCCGCTCCCGCTGCCCGACGCCACCGCGCTCGACTGGGCCGCGCTCGTGCACACCGCCACGCGCGCCATGCTGCAGATCTGCGAAGAACATCAATATCCATCAATAGATAATACTGATCCTGCTTTGGAAGCTCTCAGCACGGAACCTATAAACGATTCCTGGTCTGACGTCACACAAGTGAGTCCGCAGCCGACGGCGAGTGCGGGCGCGGGCACGCCGAGTGCGTGCGCGTGTGCGTGCGGGCTGGCGTCGCGCGTGGCGGCGCTGGAGGCGGACCACGCCGCCGCGCAACGCCACCGCCACCAGCTCGAGGACGAGGTACGTCGTCTCCGTCGGCACAACGCGAGACTGCGCGAGGAGTCTGCTCGGGCCGTGCTGCAACTGCGCCACTTCACGCAATGGTTGAAGAGGACAGTCGACAGACAATAA
Protein
MYAPDQRRKETRDVIYRSNSSLDLIYTEHSTGNGLRREYGSHGSIDVIGGANDRFPAMVHNYNGISTPEKHPGLTKANERFTEVNSDHADGPTPRSSASPKTRLKLNKLWGAASGTRPQLDEVSVSSVTDVSKRKQFAHYDCQSLMTNLNYAAKVRGELLSRRRNTTTGASAASATRGLPSGEDALGDTGDGRSNELLESCPYFRNELGGEEERCVSLCWRTARGAIIGQAGGWHRPRTSYGISMLEFPPGHSHWRNGCPFVRSMAPLPIESQDLGALYYRNYFYNQPHQNWFGMDDILGPVAVSIKKERVELRRGNTDASTPLSQLVWQYRLLIRTSELLTLRGSILEDALINIKQTNSASYNTKEVLEYVAPELQLNCLRLGISNRGAEEQLLRLDEQCVTRHYKVGVMYCKSGQSTEEEMYNNQEAGPAFVEFLQMLGQTVRLKDFDKYKAGLDNKTDSTGLYSVFSTYQSCDIMFHVSTMLPYTPNNRQQLLRKRHIGNDIVTIVFQEPGAAPFTPRNIRSQFQHVFIVVRAIDPCTDHTHYSIAVSRAKEVPLFGPPIKDGAVYPKGEAFTDLLLSKVINGENAAIQSPKFSTMATRTRQEYLKDLAKNYITATTVETGQKFSLFSSLLMKSGSNSANSTMVARVDNCTNDVLVGGALCFHVHVQDEGAGGTLVECIMGISADTIVFVEDLNRQIILVLPTASLLGFHVQPDGLVTAVEGGGCADRAGIRRGARLLEVCRAAVVALNHDQLVDLLKTSDPVTVRVTFASGLKCQCETGGEEWAAQARADAQQPARGRRAYERGHSPPRSDSSGYGTGGSGRSGRRSPDGVRRRSHDDSSPRHSRQKRSSANTSSLTEELMRLMDAELGPGRLESSAAPLPLPDATALDWAALVHTATRAMLQICEEHQYPSIDNTDPALEALSTEPINDSWSDVTQVSPQPTASAGAGTPSACACACGLASRVAALEADHAAAQRHRHQLEDEVRRLRRHNARLREESARAVLQLRHFTQWLKRTVDRQ
Summary
Uniprot
H9JLF7
A0A2A4JBB3
A0A2H1VV80
A0A2W1BWX2
A0A212F7A7
A0A194R009
+ More
A0A194PHE3 A0A0L7LG12 A0A067RTP3 E0VP23 A0A1Y1LT40 A0A023F1P8 D2A2V9 A0A139WIV1 A0A1W4WTY3 A0A1W4X3I5 A0A1W4X4V2 A0A1W4X4Q3 A0A1B6CT63 A0A1B6G8M8 A0A158NYI6 E9IUU9 A0A026WMK9 A0A182Y5J4 K7IQP6 A0A336M9N5 A0A0L7RE37 A0A154PB02 A0A1Q3F641 A0A1Q3F5P3 A0A0C9PLE2 B0W8T4 A0A182WHH3 U4URY1 A0A151WQF0 Q17IV2 A0A195AZ58 A0A182JZZ1 A0A182RRK1 A0A182M995 A0A087ZYY5 A0A195F6A5 F4X346 A0A2H8TTY3 A0A2S2NNG0 T1IPW2 A0A2S2Q3B1 A0A182X155 A0A182TQL1 A0A182NUV3 J9KA06 A0A195CWF4 A0A151J3Y8 A0A2A3EE12
A0A194PHE3 A0A0L7LG12 A0A067RTP3 E0VP23 A0A1Y1LT40 A0A023F1P8 D2A2V9 A0A139WIV1 A0A1W4WTY3 A0A1W4X3I5 A0A1W4X4V2 A0A1W4X4Q3 A0A1B6CT63 A0A1B6G8M8 A0A158NYI6 E9IUU9 A0A026WMK9 A0A182Y5J4 K7IQP6 A0A336M9N5 A0A0L7RE37 A0A154PB02 A0A1Q3F641 A0A1Q3F5P3 A0A0C9PLE2 B0W8T4 A0A182WHH3 U4URY1 A0A151WQF0 Q17IV2 A0A195AZ58 A0A182JZZ1 A0A182RRK1 A0A182M995 A0A087ZYY5 A0A195F6A5 F4X346 A0A2H8TTY3 A0A2S2NNG0 T1IPW2 A0A2S2Q3B1 A0A182X155 A0A182TQL1 A0A182NUV3 J9KA06 A0A195CWF4 A0A151J3Y8 A0A2A3EE12
Pubmed
EMBL
BABH01005844
BABH01005845
NWSH01002022
PCG69397.1
ODYU01004601
SOQ44656.1
+ More
KZ149911 PZC78124.1 AGBW02009893 OWR49624.1 KQ460883 KPJ11133.1 KQ459603 KPI92831.1 JTDY01001341 KOB74141.1 KK852424 KDR24180.1 DS235354 EEB15129.1 GEZM01047351 JAV76874.1 GBBI01003479 JAC15233.1 KQ971338 EFA01997.2 KYB27859.1 GEDC01020711 JAS16587.1 GECZ01010979 JAS58790.1 ADTU01003927 GL766050 EFZ15647.1 KK107151 QOIP01000003 EZA57312.1 RLU24550.1 UFQS01000541 UFQT01000541 SSX04724.1 SSX25087.1 KQ414613 KOC68991.1 KQ434864 KZC09089.1 GFDL01012028 JAV23017.1 GFDL01012169 JAV22876.1 GBYB01001866 GBYB01001868 JAG71633.1 JAG71635.1 DS231860 EDS39313.1 KB632418 ERL95293.1 KQ982843 KYQ50033.1 CH477236 EAT46641.1 KQ976701 KYM77320.1 AXCM01003608 KQ981768 KYN35916.1 GL888608 EGI59146.1 GFXV01005862 MBW17667.1 GGMR01006059 MBY18678.1 JH431265 GGMS01003022 MBY72225.1 ABLF02039972 KQ977220 KYN04867.1 KQ980228 KYN17149.1 KZ288269 PBC29967.1
KZ149911 PZC78124.1 AGBW02009893 OWR49624.1 KQ460883 KPJ11133.1 KQ459603 KPI92831.1 JTDY01001341 KOB74141.1 KK852424 KDR24180.1 DS235354 EEB15129.1 GEZM01047351 JAV76874.1 GBBI01003479 JAC15233.1 KQ971338 EFA01997.2 KYB27859.1 GEDC01020711 JAS16587.1 GECZ01010979 JAS58790.1 ADTU01003927 GL766050 EFZ15647.1 KK107151 QOIP01000003 EZA57312.1 RLU24550.1 UFQS01000541 UFQT01000541 SSX04724.1 SSX25087.1 KQ414613 KOC68991.1 KQ434864 KZC09089.1 GFDL01012028 JAV23017.1 GFDL01012169 JAV22876.1 GBYB01001866 GBYB01001868 JAG71633.1 JAG71635.1 DS231860 EDS39313.1 KB632418 ERL95293.1 KQ982843 KYQ50033.1 CH477236 EAT46641.1 KQ976701 KYM77320.1 AXCM01003608 KQ981768 KYN35916.1 GL888608 EGI59146.1 GFXV01005862 MBW17667.1 GGMR01006059 MBY18678.1 JH431265 GGMS01003022 MBY72225.1 ABLF02039972 KQ977220 KYN04867.1 KQ980228 KYN17149.1 KZ288269 PBC29967.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000027135 UP000009046 UP000007266 UP000192223 UP000005205 UP000053097 UP000279307 UP000076408 UP000002358 UP000053825 UP000076502 UP000002320 UP000075920 UP000030742 UP000075809 UP000008820 UP000078540 UP000075881 UP000075900 UP000075883 UP000005203 UP000078541 UP000007755 UP000076407 UP000075902 UP000075884 UP000007819 UP000078542 UP000078492 UP000242457
UP000027135 UP000009046 UP000007266 UP000192223 UP000005205 UP000053097 UP000279307 UP000076408 UP000002358 UP000053825 UP000076502 UP000002320 UP000075920 UP000030742 UP000075809 UP000008820 UP000078540 UP000075881 UP000075900 UP000075883 UP000005203 UP000078541 UP000007755 UP000076407 UP000075902 UP000075884 UP000007819 UP000078542 UP000078492 UP000242457
PRIDE
Gene 3D
ProteinModelPortal
H9JLF7
A0A2A4JBB3
A0A2H1VV80
A0A2W1BWX2
A0A212F7A7
A0A194R009
+ More
A0A194PHE3 A0A0L7LG12 A0A067RTP3 E0VP23 A0A1Y1LT40 A0A023F1P8 D2A2V9 A0A139WIV1 A0A1W4WTY3 A0A1W4X3I5 A0A1W4X4V2 A0A1W4X4Q3 A0A1B6CT63 A0A1B6G8M8 A0A158NYI6 E9IUU9 A0A026WMK9 A0A182Y5J4 K7IQP6 A0A336M9N5 A0A0L7RE37 A0A154PB02 A0A1Q3F641 A0A1Q3F5P3 A0A0C9PLE2 B0W8T4 A0A182WHH3 U4URY1 A0A151WQF0 Q17IV2 A0A195AZ58 A0A182JZZ1 A0A182RRK1 A0A182M995 A0A087ZYY5 A0A195F6A5 F4X346 A0A2H8TTY3 A0A2S2NNG0 T1IPW2 A0A2S2Q3B1 A0A182X155 A0A182TQL1 A0A182NUV3 J9KA06 A0A195CWF4 A0A151J3Y8 A0A2A3EE12
A0A194PHE3 A0A0L7LG12 A0A067RTP3 E0VP23 A0A1Y1LT40 A0A023F1P8 D2A2V9 A0A139WIV1 A0A1W4WTY3 A0A1W4X3I5 A0A1W4X4V2 A0A1W4X4Q3 A0A1B6CT63 A0A1B6G8M8 A0A158NYI6 E9IUU9 A0A026WMK9 A0A182Y5J4 K7IQP6 A0A336M9N5 A0A0L7RE37 A0A154PB02 A0A1Q3F641 A0A1Q3F5P3 A0A0C9PLE2 B0W8T4 A0A182WHH3 U4URY1 A0A151WQF0 Q17IV2 A0A195AZ58 A0A182JZZ1 A0A182RRK1 A0A182M995 A0A087ZYY5 A0A195F6A5 F4X346 A0A2H8TTY3 A0A2S2NNG0 T1IPW2 A0A2S2Q3B1 A0A182X155 A0A182TQL1 A0A182NUV3 J9KA06 A0A195CWF4 A0A151J3Y8 A0A2A3EE12
PDB
1SRQ
E-value=4.83725e-55,
Score=547
Ontologies
GO
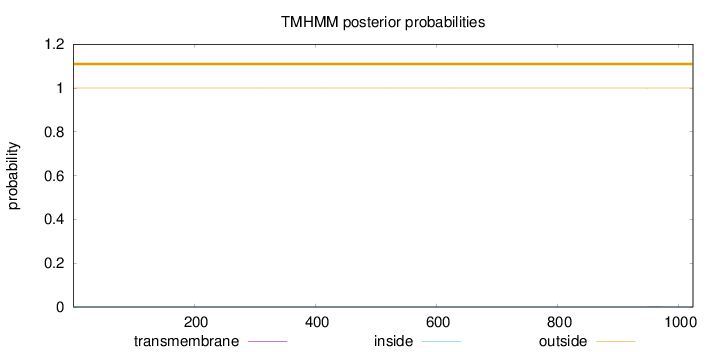
Topology
Length:
1024
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00815
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00009
outside
1 - 1024
Population Genetic Test Statistics
Pi
17.240531
Theta
15.708334
Tajima's D
0.323002
CLR
0.575279
CSRT
0.475776211189441
Interpretation
Uncertain
