Gene
KWMTBOMO07250 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010361
Annotation
muscle_glycogen_phosphorylase_[Bombyx_mori]
Full name
Alpha-1,4 glucan phosphorylase
+ More
Multifunctional fusion protein
Glycogen phosphorylase
Multifunctional fusion protein
Glycogen phosphorylase
Location in the cell
Cytoplasmic Reliability : 3.051
Sequence
CDS
ATGAGCGTACAGACTGACGCGGAAAAGCGTAAACAGATATCTGTCAGAGGAATTGTTGCTGTCGAAAATGTGACTGAAGTAAAGAAGGCGTTCAATCGTCATGTCCATTACACTTTAGTGAAAGATCGAAATGTTGCGACCCCGCGGGATTACTATTTTGCTCTCGCACACACCGTGAAAGATCATCTTGTCTCCAGGTGGATTCGTACTCAACAATACTATTATGAAAATGATCCTAAGAGAGTCTACTATTTGTCCTTGGAGTACTATATGGGGCGTTCTCTTCAAAATACCATGATCAACCTTGGCATCCAAGGTACTGTTGACGAAGCATTATACCAACTTGGTCTGGATATTGAAGAATTAGAAGAACTTGAGGAAGATGCTGGTCTGGGAAATGGAGGTCTTGGACGCTTAGCAGCTTGTTTCCTAGACTCTATGGCGACATTAGGTCTTGCTGCGTATGGATATGGTATAAGGTACGAATATGGTATATTTGCTCAAAAAATTGAAAACGGAGAACAACAGGAAGAGCCTGATGATTGGTTACGTTTCGGAAACCCCTGGGAGAAGGCAAGACCTGAGTTCATGCTGCCAGTTAACTTCTATGGTAGTGTAGTTGACACACCCGATGGAAAAAAATGGATTGATACACAGGTGGTATTTGCAATGCCATACGATAACCCTATTCCTGGTTATAACAATAATGTTGTGAATACATTGCGACTGTGGTCGGCGAAGAGCCCCATTGATTTCAATCTTAAATTTTTCAATTCTGGTGATTATATTCAAGCTGTATTGGATCGCAACGTTGCCGAAAACATATCGCGTGTTTTGTATCCCAACGACAACTTCTTCGAAGGCAAAGAGTTGCGTTTGCGCCAAGAATACTTCATGTGTGCTGCCACGCTCCAGGATATTATCAGACGGTACAAAGCATCTAAGTTCGGATGCCGAGATGCCGTTAGAACTTCGTTTGAACATTTACCTGAAAAAGTAGCCATCCAATTAAATGACACTCATCCGGCTCTGGCTATACCTGAATTTTTGAGAATCTTAGTCGACATTGAAAAGGTTCCCTATGAACAAGCTTGGGACTTAGTTGTTAAATGTTGTGCATACACCAACCACACAGTCCTGCCTGAGGCTCTCGAAAGATGGCCATGCTCGATGCTGGAAAATGTTCTGCCGAGACATATGCAATTGATTTACCATATCAACTTTTTACATTTGCAAGAGGTGCAAAAGCGTTGGCCCGGCGACATGGATCGTTTGCGCAGAATGTCCTTGATTGAAGAAGAAGGTGAAAAACGCGTCAATATGGCTAATTTATCTATTGTCGGCTCTCACGCTGTCAATGGTGTAGCAGCCATTCACTCCGAAATCTTAAAAGCCACTATTTTCCGCGACTTTTTCGAAATGTGGCCTGATAAATTCCAAAATAAGACTAACGGCATCACTCCCCGGCGTTGGTTGCTCCTCTGCAACCCTGGCTTATCTGATCTCATCTGCGACAAGATCGGAGAAGATTGGATCGTCCATTTGGAGAAGTTGAAGGAGCTGAAGCGCTGGGCTAAGGACCCGGCTTTCCAGCGCGCCGTTATGAAAGTGAAGCAAGAGAATAAACTCAAGCTGGCGGCCCTTATTGAAAGGGATACCGGAGTTAAAATTAATCCCGCGTCGATGTTCGATGTCCAAGTGAAGCGCATCCACGAGTACAAACGTCAGTTACTGAATATTTTGCACGTGATCACGCTTTACAACCGCATCAAGCGCGACCCCAGCGCGTCTTTCACGCCGAGGACTGTTATGATCGGTGGCAAAGCCGCTCCTGGATACTTCATCGCCAAACAAATTATCGCTTTAGCCTGTGCAGTTGGAAATACTGTGAACAACGACCCAGACGTAGGTGACAAGTTGAAGCTGATATTCCTCGAGAACTATCGGGTGACCTTGGCAGAGCGTATCATCCCGGCATCGGATTTGAGCGAACAGATTTCAACAGCAGGAACTGAGGCATCCGGCACCGGCAATATGAAGTTTATGCTAAATGGAGCGTTAACTATTGGCACCATGGACGGAGCCAACGTTGAAATGGCCGAAGAAGCTGGTGAAGAGAACTTTTTCATTTTCGGAATGAGAGTTGACGATGTCGAAGCACTGAAACAGCGCGGGTACAACGCATACGAGTACTACGAGCGCAACCCGGAGCTGCGGCAGTGCGTGGAGCAGATCCGCAGCGGGTTCTTCTCGACGGGCGAGCCCGGCAAGTTCGCGCACGTCGCCGACGTGCTGCTGCACCACGACCGCTTCCTGCACCTCGCCGACTACGACGCTTACGTCGAGGCCCAGCAGAAAGTCGCCGACGTATACCAGGACCAGACAAAATGGGCGGAGATGGTGATCGAGAACATCGCGTCGTCTGGCAAGTTCTCGTCTGACCGCACCATCACGGAGTACGCCCGCGAAATCTGGGGCGTGGAGCCCACCTGGGACAAGCTGCCCGCCCCGCACGAGACCGCTTAG
Protein
MSVQTDAEKRKQISVRGIVAVENVTEVKKAFNRHVHYTLVKDRNVATPRDYYFALAHTVKDHLVSRWIRTQQYYYENDPKRVYYLSLEYYMGRSLQNTMINLGIQGTVDEALYQLGLDIEELEELEEDAGLGNGGLGRLAACFLDSMATLGLAAYGYGIRYEYGIFAQKIENGEQQEEPDDWLRFGNPWEKARPEFMLPVNFYGSVVDTPDGKKWIDTQVVFAMPYDNPIPGYNNNVVNTLRLWSAKSPIDFNLKFFNSGDYIQAVLDRNVAENISRVLYPNDNFFEGKELRLRQEYFMCAATLQDIIRRYKASKFGCRDAVRTSFEHLPEKVAIQLNDTHPALAIPEFLRILVDIEKVPYEQAWDLVVKCCAYTNHTVLPEALERWPCSMLENVLPRHMQLIYHINFLHLQEVQKRWPGDMDRLRRMSLIEEEGEKRVNMANLSIVGSHAVNGVAAIHSEILKATIFRDFFEMWPDKFQNKTNGITPRRWLLLCNPGLSDLICDKIGEDWIVHLEKLKELKRWAKDPAFQRAVMKVKQENKLKLAALIERDTGVKINPASMFDVQVKRIHEYKRQLLNILHVITLYNRIKRDPSASFTPRTVMIGGKAAPGYFIAKQIIALACAVGNTVNNDPDVGDKLKLIFLENYRVTLAERIIPASDLSEQISTAGTEASGTGNMKFMLNGALTIGTMDGANVEMAEEAGEENFFIFGMRVDDVEALKQRGYNAYEYYERNPELRQCVEQIRSGFFSTGEPGKFAHVADVLLHHDRFLHLADYDAYVEAQQKVADVYQDQTKWAEMVIENIASSGKFSSDRTITEYAREIWGVEPTWDKLPAPHETA
Summary
Description
Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties.
Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties (By similarity). Required for glycogen breakdown in skeletal muscle.
Phosphorylase is an important allosteric enzyme in carbohydrate metabolism. Enzymes from different sources differ in their regulatory mechanisms and in their natural substrates. However, all known phosphorylases share catalytic and structural properties (By similarity). Required for glycogen breakdown in skeletal muscle.
Catalytic Activity
[(1->4)-alpha-D-glucosyl](n) + phosphate = [(1->4)-alpha-D-glucosyl](n-1) + alpha-D-glucose 1-phosphate
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the glycogen phosphorylase family.
Keywords
Carbohydrate metabolism
Complete proteome
Glycogen metabolism
Glycosyltransferase
Phosphoprotein
Pyridoxal phosphate
Reference proteome
Transferase
Feature
chain Glycogen phosphorylase
Uniprot
B2CMB5
H9JLG0
A0A2H1VEE6
A0A2W1BUS8
A0A2A4JHR0
A0A194R5J6
+ More
S4PAM0 C0LRA9 A0A194PIF2 A0A2S1TNQ4 I7CKW1 A0A182L220 A0A182F4K3 A0A0L0BUC4 A0A182W9P0 A0A182TUX3 A0A084VP10 A0A3Q8B932 A0A2M4A0E8 A0A2M3YY01 A0A2Y9D354 A0A2Y9D126 Q7Q3L6 A0A182WT70 A0A1Y9H2J4 A0A2A3E7B2 W5JD07 A0A182MV21 A0A2M4BDP3 A0A336K387 A0A087ZVF4 A0A182QG57 A0A182YGT4 A0A182RBK7 B4KI29 A0A182JPV9 U5EZ80 B3N9F7 A4UZZ4 A0A310SEB9 Q17NG8 Q9XTL9 A0A182HDR9 A0A182J4Q7 A0A023EVY7 B3MJC8 T1PGX7 A0A0J9QU28 A0A1W4UTM2 B4LTC6 A0A1I8NIE9 B4NW47 B4MVT1 A0A154PP88 D2A0P2 B0WCF2 A0A0M4EN07 B4JAB0 A0A1L8E1Y2 A0A1Q3FM82 A0A1I8PPB2 A0A0K8VMW2 A0A026VYY3 A0A0K8UVG6 A0A0L7QRZ2 A0A3B0K2R5 A0A0T6BFY4 A0A034WBQ5 K7IQK0 A0A232F127 L7X9Z5 W8ARZ5 Q29MI1 B4G970 A0A0A1X1Y6 A0A0A9Y0L7 A0A1A9UT61 A0A0A9XZ40 A0A1W5YLJ9 E2BWP7 A0A0M9AA77 A0A0C9RJQ2 A0A158NVR8 A0A195EC44 E1ZZF6 A0A1Y1M6I5 A0A195B8I0 A0A286SDL8 F4WNR3 A0A346A6L6 A0A1A9WUG4 E2S085 A0A224X783 A0A023F5H7 T1I3T6 A0A2P1JJ59 U4UTB8 A0A087U449 E9G2G6 J9WM54
S4PAM0 C0LRA9 A0A194PIF2 A0A2S1TNQ4 I7CKW1 A0A182L220 A0A182F4K3 A0A0L0BUC4 A0A182W9P0 A0A182TUX3 A0A084VP10 A0A3Q8B932 A0A2M4A0E8 A0A2M3YY01 A0A2Y9D354 A0A2Y9D126 Q7Q3L6 A0A182WT70 A0A1Y9H2J4 A0A2A3E7B2 W5JD07 A0A182MV21 A0A2M4BDP3 A0A336K387 A0A087ZVF4 A0A182QG57 A0A182YGT4 A0A182RBK7 B4KI29 A0A182JPV9 U5EZ80 B3N9F7 A4UZZ4 A0A310SEB9 Q17NG8 Q9XTL9 A0A182HDR9 A0A182J4Q7 A0A023EVY7 B3MJC8 T1PGX7 A0A0J9QU28 A0A1W4UTM2 B4LTC6 A0A1I8NIE9 B4NW47 B4MVT1 A0A154PP88 D2A0P2 B0WCF2 A0A0M4EN07 B4JAB0 A0A1L8E1Y2 A0A1Q3FM82 A0A1I8PPB2 A0A0K8VMW2 A0A026VYY3 A0A0K8UVG6 A0A0L7QRZ2 A0A3B0K2R5 A0A0T6BFY4 A0A034WBQ5 K7IQK0 A0A232F127 L7X9Z5 W8ARZ5 Q29MI1 B4G970 A0A0A1X1Y6 A0A0A9Y0L7 A0A1A9UT61 A0A0A9XZ40 A0A1W5YLJ9 E2BWP7 A0A0M9AA77 A0A0C9RJQ2 A0A158NVR8 A0A195EC44 E1ZZF6 A0A1Y1M6I5 A0A195B8I0 A0A286SDL8 F4WNR3 A0A346A6L6 A0A1A9WUG4 E2S085 A0A224X783 A0A023F5H7 T1I3T6 A0A2P1JJ59 U4UTB8 A0A087U449 E9G2G6 J9WM54
EC Number
2.4.1.1
Pubmed
19121390
28756777
26354079
23622113
20966253
26108605
+ More
24438588 12364791 14747013 17210077 20920257 23761445 25244985 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17510324 10092506 10731138 24265594 26483478 24945155 22936249 25315136 17550304 18362917 19820115 24508170 30249741 25348373 20075255 28648823 24495485 15632085 25830018 25401762 20798317 21347285 28004739 21719571 29187976 25474469 23537049 21292972 22972362
24438588 12364791 14747013 17210077 20920257 23761445 25244985 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17510324 10092506 10731138 24265594 26483478 24945155 22936249 25315136 17550304 18362917 19820115 24508170 30249741 25348373 20075255 28648823 24495485 15632085 25830018 25401762 20798317 21347285 28004739 21719571 29187976 25474469 23537049 21292972 22972362
EMBL
EU527367
ACB41088.1
BABH01005840
BABH01005841
BABH01005842
ODYU01002089
+ More
SOQ39156.1 KZ149911 PZC78121.1 NWSH01001519 PCG70970.1 KQ460883 KPJ11136.1 GAIX01004946 JAA87614.1 FJ754277 ACN78408.1 KQ459603 KPI92828.1 MG517442 AWI63366.1 JX113672 AFO54708.2 JRES01001454 KNC22809.1 ATLV01014966 KE524999 KFB39704.1 KU556838 AQT25653.1 GGFK01000908 MBW34229.1 GGFM01000365 MBW21116.1 APCN01002494 AB523399 AAAB01008964 BAI60046.1 EAA12902.3 KZ288345 PBC27637.1 ADMH02001514 ETN62232.1 AXCM01007187 AXCM01007188 GGFJ01002021 MBW51162.1 UFQS01000097 UFQT01000097 SSW99426.1 SSX19806.1 AXCN02000872 CH933807 EDW11311.1 GANO01001273 JAB58598.1 CH954177 EDV57414.1 AE014134 AAZ66442.1 KQ762207 OAD56035.1 CH477199 EAT48250.1 AF073177 AF073178 AF073179 AF160947 JXUM01034765 JXUM01034766 KQ561035 KXJ79955.1 JXUM01055500 JXUM01055501 JXUM01055502 GAPW01000210 KQ561870 JAC13388.1 KXJ77284.1 CH902620 EDV31338.1 KA647395 AFP62024.1 CM002910 KMY87582.1 CH940649 EDW63896.1 CM000157 EDW87327.1 CH963857 EDW75801.1 KQ435007 KZC13705.1 KQ971338 EFA02541.1 DS231887 EDS43476.1 CP012523 ALC37975.1 CH916368 EDW03781.1 GFDF01001366 JAV12718.1 GFDL01006427 JAV28618.1 GDHF01012404 JAI39910.1 KK107558 QOIP01000012 EZA48982.1 RLU15903.1 GDHF01021989 JAI30325.1 KQ414766 KOC61387.1 OUUW01000004 SPP79301.1 LJIG01000676 KRT86264.1 GAKP01006823 JAC52129.1 NNAY01001358 OXU24250.1 JX974433 AGC97435.1 GAMC01019202 JAB87353.1 CH379060 EAL33713.1 CH479180 EDW28900.1 GBXI01009609 JAD04683.1 GBHO01018423 JAG25181.1 GBHO01018420 JAG25184.1 KX147656 ARI45060.1 GL451165 EFN79902.1 KQ435701 KOX80377.1 GBYB01013502 JAG83269.1 ADTU01002901 ADTU01002902 KQ979074 KYN22795.1 GL435343 EFN73448.1 GEZM01039845 JAV81141.1 KQ976565 KYM80547.1 KX766454 ASZ80181.1 GL888237 EGI64256.1 MF908516 AXK92638.1 AB596876 BAJ23879.1 GFTR01008160 JAW08266.1 GBBI01002304 JAC16408.1 ACPB03003299 MF385058 AVN99053.1 KB632354 ERL93360.1 KK118066 KFM72138.1 GL732530 EFX86312.1 JX462658 AFS17314.1
SOQ39156.1 KZ149911 PZC78121.1 NWSH01001519 PCG70970.1 KQ460883 KPJ11136.1 GAIX01004946 JAA87614.1 FJ754277 ACN78408.1 KQ459603 KPI92828.1 MG517442 AWI63366.1 JX113672 AFO54708.2 JRES01001454 KNC22809.1 ATLV01014966 KE524999 KFB39704.1 KU556838 AQT25653.1 GGFK01000908 MBW34229.1 GGFM01000365 MBW21116.1 APCN01002494 AB523399 AAAB01008964 BAI60046.1 EAA12902.3 KZ288345 PBC27637.1 ADMH02001514 ETN62232.1 AXCM01007187 AXCM01007188 GGFJ01002021 MBW51162.1 UFQS01000097 UFQT01000097 SSW99426.1 SSX19806.1 AXCN02000872 CH933807 EDW11311.1 GANO01001273 JAB58598.1 CH954177 EDV57414.1 AE014134 AAZ66442.1 KQ762207 OAD56035.1 CH477199 EAT48250.1 AF073177 AF073178 AF073179 AF160947 JXUM01034765 JXUM01034766 KQ561035 KXJ79955.1 JXUM01055500 JXUM01055501 JXUM01055502 GAPW01000210 KQ561870 JAC13388.1 KXJ77284.1 CH902620 EDV31338.1 KA647395 AFP62024.1 CM002910 KMY87582.1 CH940649 EDW63896.1 CM000157 EDW87327.1 CH963857 EDW75801.1 KQ435007 KZC13705.1 KQ971338 EFA02541.1 DS231887 EDS43476.1 CP012523 ALC37975.1 CH916368 EDW03781.1 GFDF01001366 JAV12718.1 GFDL01006427 JAV28618.1 GDHF01012404 JAI39910.1 KK107558 QOIP01000012 EZA48982.1 RLU15903.1 GDHF01021989 JAI30325.1 KQ414766 KOC61387.1 OUUW01000004 SPP79301.1 LJIG01000676 KRT86264.1 GAKP01006823 JAC52129.1 NNAY01001358 OXU24250.1 JX974433 AGC97435.1 GAMC01019202 JAB87353.1 CH379060 EAL33713.1 CH479180 EDW28900.1 GBXI01009609 JAD04683.1 GBHO01018423 JAG25181.1 GBHO01018420 JAG25184.1 KX147656 ARI45060.1 GL451165 EFN79902.1 KQ435701 KOX80377.1 GBYB01013502 JAG83269.1 ADTU01002901 ADTU01002902 KQ979074 KYN22795.1 GL435343 EFN73448.1 GEZM01039845 JAV81141.1 KQ976565 KYM80547.1 KX766454 ASZ80181.1 GL888237 EGI64256.1 MF908516 AXK92638.1 AB596876 BAJ23879.1 GFTR01008160 JAW08266.1 GBBI01002304 JAC16408.1 ACPB03003299 MF385058 AVN99053.1 KB632354 ERL93360.1 KK118066 KFM72138.1 GL732530 EFX86312.1 JX462658 AFS17314.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000075882
UP000069272
+ More
UP000037069 UP000075920 UP000075902 UP000030765 UP000075903 UP000075840 UP000007062 UP000076407 UP000075884 UP000242457 UP000000673 UP000075883 UP000005203 UP000075886 UP000076408 UP000075900 UP000009192 UP000075881 UP000008711 UP000000803 UP000008820 UP000069940 UP000249989 UP000075880 UP000007801 UP000192221 UP000008792 UP000095301 UP000002282 UP000007798 UP000076502 UP000007266 UP000002320 UP000092553 UP000001070 UP000095300 UP000053097 UP000279307 UP000053825 UP000268350 UP000002358 UP000215335 UP000001819 UP000008744 UP000078200 UP000008237 UP000053105 UP000005205 UP000078492 UP000000311 UP000078540 UP000007755 UP000091820 UP000015103 UP000030742 UP000054359 UP000000305
UP000037069 UP000075920 UP000075902 UP000030765 UP000075903 UP000075840 UP000007062 UP000076407 UP000075884 UP000242457 UP000000673 UP000075883 UP000005203 UP000075886 UP000076408 UP000075900 UP000009192 UP000075881 UP000008711 UP000000803 UP000008820 UP000069940 UP000249989 UP000075880 UP000007801 UP000192221 UP000008792 UP000095301 UP000002282 UP000007798 UP000076502 UP000007266 UP000002320 UP000092553 UP000001070 UP000095300 UP000053097 UP000279307 UP000053825 UP000268350 UP000002358 UP000215335 UP000001819 UP000008744 UP000078200 UP000008237 UP000053105 UP000005205 UP000078492 UP000000311 UP000078540 UP000007755 UP000091820 UP000015103 UP000030742 UP000054359 UP000000305
Interpro
ProteinModelPortal
B2CMB5
H9JLG0
A0A2H1VEE6
A0A2W1BUS8
A0A2A4JHR0
A0A194R5J6
+ More
S4PAM0 C0LRA9 A0A194PIF2 A0A2S1TNQ4 I7CKW1 A0A182L220 A0A182F4K3 A0A0L0BUC4 A0A182W9P0 A0A182TUX3 A0A084VP10 A0A3Q8B932 A0A2M4A0E8 A0A2M3YY01 A0A2Y9D354 A0A2Y9D126 Q7Q3L6 A0A182WT70 A0A1Y9H2J4 A0A2A3E7B2 W5JD07 A0A182MV21 A0A2M4BDP3 A0A336K387 A0A087ZVF4 A0A182QG57 A0A182YGT4 A0A182RBK7 B4KI29 A0A182JPV9 U5EZ80 B3N9F7 A4UZZ4 A0A310SEB9 Q17NG8 Q9XTL9 A0A182HDR9 A0A182J4Q7 A0A023EVY7 B3MJC8 T1PGX7 A0A0J9QU28 A0A1W4UTM2 B4LTC6 A0A1I8NIE9 B4NW47 B4MVT1 A0A154PP88 D2A0P2 B0WCF2 A0A0M4EN07 B4JAB0 A0A1L8E1Y2 A0A1Q3FM82 A0A1I8PPB2 A0A0K8VMW2 A0A026VYY3 A0A0K8UVG6 A0A0L7QRZ2 A0A3B0K2R5 A0A0T6BFY4 A0A034WBQ5 K7IQK0 A0A232F127 L7X9Z5 W8ARZ5 Q29MI1 B4G970 A0A0A1X1Y6 A0A0A9Y0L7 A0A1A9UT61 A0A0A9XZ40 A0A1W5YLJ9 E2BWP7 A0A0M9AA77 A0A0C9RJQ2 A0A158NVR8 A0A195EC44 E1ZZF6 A0A1Y1M6I5 A0A195B8I0 A0A286SDL8 F4WNR3 A0A346A6L6 A0A1A9WUG4 E2S085 A0A224X783 A0A023F5H7 T1I3T6 A0A2P1JJ59 U4UTB8 A0A087U449 E9G2G6 J9WM54
S4PAM0 C0LRA9 A0A194PIF2 A0A2S1TNQ4 I7CKW1 A0A182L220 A0A182F4K3 A0A0L0BUC4 A0A182W9P0 A0A182TUX3 A0A084VP10 A0A3Q8B932 A0A2M4A0E8 A0A2M3YY01 A0A2Y9D354 A0A2Y9D126 Q7Q3L6 A0A182WT70 A0A1Y9H2J4 A0A2A3E7B2 W5JD07 A0A182MV21 A0A2M4BDP3 A0A336K387 A0A087ZVF4 A0A182QG57 A0A182YGT4 A0A182RBK7 B4KI29 A0A182JPV9 U5EZ80 B3N9F7 A4UZZ4 A0A310SEB9 Q17NG8 Q9XTL9 A0A182HDR9 A0A182J4Q7 A0A023EVY7 B3MJC8 T1PGX7 A0A0J9QU28 A0A1W4UTM2 B4LTC6 A0A1I8NIE9 B4NW47 B4MVT1 A0A154PP88 D2A0P2 B0WCF2 A0A0M4EN07 B4JAB0 A0A1L8E1Y2 A0A1Q3FM82 A0A1I8PPB2 A0A0K8VMW2 A0A026VYY3 A0A0K8UVG6 A0A0L7QRZ2 A0A3B0K2R5 A0A0T6BFY4 A0A034WBQ5 K7IQK0 A0A232F127 L7X9Z5 W8ARZ5 Q29MI1 B4G970 A0A0A1X1Y6 A0A0A9Y0L7 A0A1A9UT61 A0A0A9XZ40 A0A1W5YLJ9 E2BWP7 A0A0M9AA77 A0A0C9RJQ2 A0A158NVR8 A0A195EC44 E1ZZF6 A0A1Y1M6I5 A0A195B8I0 A0A286SDL8 F4WNR3 A0A346A6L6 A0A1A9WUG4 E2S085 A0A224X783 A0A023F5H7 T1I3T6 A0A2P1JJ59 U4UTB8 A0A087U449 E9G2G6 J9WM54
PDB
1Z8D
E-value=0,
Score=3266
Ontologies
PATHWAY
GO
PANTHER
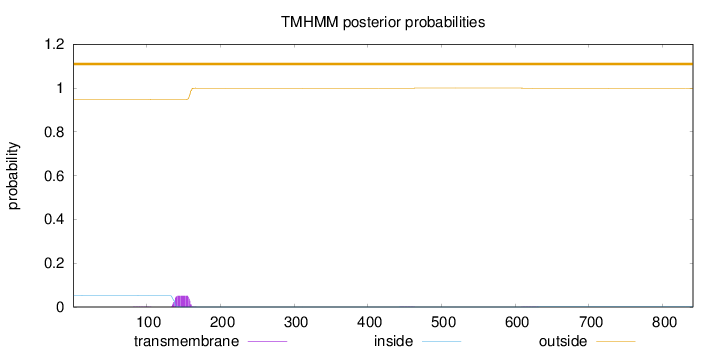
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
841
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.14493
Exp number, first 60 AAs:
0.00093
Total prob of N-in:
0.05197
outside
1 - 841
Population Genetic Test Statistics
Pi
0.721463
Theta
4.294427
Tajima's D
-2.280168
CLR
590.076746
CSRT
0.00164991750412479
Interpretation
Possibly Positive selection
