Gene
KWMTBOMO07247 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010588
Annotation
H+_transporting_ATP_synthase_delta_subunit_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.791
Sequence
CDS
ATGGCACTGGCATTCCGTGGTGTAATCAGAAATGTAGCTCGTCGGCTTCAAGTACGTAACTATGCAGATGCTCCCAAAGGTGACGAAATGGCATTGACCTTCGCTGCGGGTAATAAGGTCTTTTATGACAAACAAGTTGTCAAACAAATTGATGTACCATCATTTAGTGGTGCTTTTGGTATCTTACCTAAACACGTACCTACCCTGGCTGTTCTTAGACCTGGTGTTGTTACAATTCTTGAAAATGATGGGAAACAGAATAAAATCTTCGTATCGTCTGGCACAATTACAGTAAATGATGACTCCTCAGTTCAGGTTTTAGCCGAAGAGGCTCACCCTCTGGAGAGCATTGACCGTAGTGCTGCACAGGAGGCCCTCAGTAAAGCACAGTCTGAATTCAATTCTGCTTCTAATGAAAAGGCTAAAGCTGAAGCTGCTATTGCTCTAGAAGTAGCTGAAGAGATTGTGAAGGCTGCATCTGCGTAA
Protein
MALAFRGVIRNVARRLQVRNYADAPKGDEMALTFAAGNKVFYDKQVVKQIDVPSFSGAFGILPKHVPTLAVLRPGVVTILENDGKQNKIFVSSGTITVNDDSSVQVLAEEAHPLESIDRSAAQEALSKAQSEFNSASNEKAKAEAAIALEVAEEIVKAASA
Summary
Uniprot
Q1HQ34
A0A194R1M3
A0A2H1VA11
I4DN05
I4DJ87
Q5MGP7
+ More
A0A2W1BST9 A0A2A4JHC9 A0A194PIE8 A0A3S2P226 S4NMX1 A0A336KXP7 A0A232EQS0 D6WNL2 E9I937 V5I9G1 A0A088AHV3 A0A2A3EA88 V9IKG8 A0A0K8TPE6 A0A151I243 A0A182JD00 A0A154NZJ1 A0A182W9Y8 A0A158NG24 A0A1J1HSS8 A0A182JTW4 A0A182QUP3 A0A1B6H198 A0A0J7NQ69 A0A1B6FZI8 A0A1B6JAZ1 A0A182P0H6 A0A182IEL1 A0A1S4GWD4 A0A182V6J7 A0A182UGZ6 A0A182WTI0 A0A182L2D6 Q7PNG6 J3JW62 A0A151XBC9 A0A310SBU4 A0A026VYH1 A0A026WIB6 A0A1Q3FJD4 A0A3Q0J2R9 A0A0M9A6D1 A0A182YKW5 A0A182SNK5 A0A1S3D4R1 A0A182R3B2 A0A2J7PWH3 B0WYE7 E2BMY9 A0A195F0R3 K7J447 A0A0P4VUG0 R4G8C3 A0A195E713 A0A1B0CAD3 A0A224XYC0 A0A0V0G4B2 A0A1L8E0M3 A0A067RCX9 E2A9J2 A0A1W4XKK8 A0A1B0EXQ7 A0A2P8Z2L9 A0A2S2Q0V8 A0A023F868 F4WTL2 A0A2M4A883 A0A1A9X3A7 A0A1B6EB44 A0A1B6C446 A0A161MDX5 A0A2M4C1Z6 A0A0A1XJK8 D1FPL2 B3MZL6 A0A1Y1MM15 A0A0M3QYZ4 U4U0R7 T1E770 A0A2M4C1Z3 A0A2M3Z256 A0A3B0JCU5 W8BUU2 W5JXC4 C4WSI1 A0A182FJU0 J9K2M7 B4PXG5 A0A2H8U0K5 A0A2S2P3X4 A0A1W4VV65
A0A2W1BST9 A0A2A4JHC9 A0A194PIE8 A0A3S2P226 S4NMX1 A0A336KXP7 A0A232EQS0 D6WNL2 E9I937 V5I9G1 A0A088AHV3 A0A2A3EA88 V9IKG8 A0A0K8TPE6 A0A151I243 A0A182JD00 A0A154NZJ1 A0A182W9Y8 A0A158NG24 A0A1J1HSS8 A0A182JTW4 A0A182QUP3 A0A1B6H198 A0A0J7NQ69 A0A1B6FZI8 A0A1B6JAZ1 A0A182P0H6 A0A182IEL1 A0A1S4GWD4 A0A182V6J7 A0A182UGZ6 A0A182WTI0 A0A182L2D6 Q7PNG6 J3JW62 A0A151XBC9 A0A310SBU4 A0A026VYH1 A0A026WIB6 A0A1Q3FJD4 A0A3Q0J2R9 A0A0M9A6D1 A0A182YKW5 A0A182SNK5 A0A1S3D4R1 A0A182R3B2 A0A2J7PWH3 B0WYE7 E2BMY9 A0A195F0R3 K7J447 A0A0P4VUG0 R4G8C3 A0A195E713 A0A1B0CAD3 A0A224XYC0 A0A0V0G4B2 A0A1L8E0M3 A0A067RCX9 E2A9J2 A0A1W4XKK8 A0A1B0EXQ7 A0A2P8Z2L9 A0A2S2Q0V8 A0A023F868 F4WTL2 A0A2M4A883 A0A1A9X3A7 A0A1B6EB44 A0A1B6C446 A0A161MDX5 A0A2M4C1Z6 A0A0A1XJK8 D1FPL2 B3MZL6 A0A1Y1MM15 A0A0M3QYZ4 U4U0R7 T1E770 A0A2M4C1Z3 A0A2M3Z256 A0A3B0JCU5 W8BUU2 W5JXC4 C4WSI1 A0A182FJU0 J9K2M7 B4PXG5 A0A2H8U0K5 A0A2S2P3X4 A0A1W4VV65
Pubmed
19121390
26354079
22651552
16023793
28756777
23622113
+ More
28648823 18362917 19820115 21282665 26369729 21347285 12364791 20966253 22516182 23537049 24508170 30249741 25244985 20798317 20075255 27129103 24845553 29403074 25474469 21719571 25830018 20441151 17994087 28004739 24495485 20920257 23761445 17550304
28648823 18362917 19820115 21282665 26369729 21347285 12364791 20966253 22516182 23537049 24508170 30249741 25244985 20798317 20075255 27129103 24845553 29403074 25474469 21719571 25830018 20441151 17994087 28004739 24495485 20920257 23761445 17550304
EMBL
BABH01005835
DQ443218
ABF51307.1
KQ460883
KPJ11140.1
ODYU01001259
+ More
SOQ37222.1 AK402673 BAM19295.1 AK401355 BAM17977.1 AY829739 AAV91353.1 KZ149911 PZC78119.1 NWSH01001519 PCG70994.1 KQ459603 KPI92823.1 RSAL01000001 RVE55132.1 GAIX01014191 JAA78369.1 UFQS01001175 UFQS01001201 UFQT01001175 UFQT01001201 SSX09420.1 SSX09703.1 SSX29321.1 NNAY01002733 OXU20667.1 KQ971342 EFA04639.1 GL761674 EFZ22922.1 GALX01002995 JAB65471.1 KZ288310 PBC28610.1 JR048944 AEY60809.1 GDAI01001341 JAI16262.1 KQ976580 KYM79924.1 KQ434786 KZC05085.1 ADTU01014940 CVRI01000020 CRK91128.1 AXCN02000610 GECZ01001342 JAS68427.1 LBMM01002631 KMQ94565.1 GECZ01014164 JAS55605.1 GECU01011513 JAS96193.1 APCN01005209 AAAB01008964 EAA12214.5 APGK01016340 APGK01026055 APGK01026056 BT127480 KB740613 KB739824 AEE62442.1 ENN79996.1 ENN82108.1 KQ982335 KYQ57629.1 KQ794714 OAD47026.1 KK107750 QOIP01000012 EZA47909.1 RLU16007.1 KK107185 EZA55792.1 GFDL01007407 JAV27638.1 KQ435726 KOX78160.1 NEVH01020933 PNF20683.1 DS232187 EDS37024.1 GL449382 EFN82893.1 KQ981864 KYN34170.1 AAZX01007151 GDKW01000413 JAI56182.1 ACPB03021503 GAHY01001222 JAA76288.1 KQ979568 KYN20978.1 AJWK01003674 GFTR01002854 JAW13572.1 GECL01003249 JAP02875.1 GFDF01001791 JAV12293.1 KK852584 KDR20848.1 GL437917 EFN69882.1 AJVK01033225 PYGN01000224 PSN50752.1 GGMS01002205 MBY71408.1 GBBI01001201 JAC17511.1 GL888341 EGI62433.1 GGFK01003686 MBW37007.1 GEDC01022451 GEDC01002132 JAS14847.1 JAS35166.1 GEDC01029134 JAS08164.1 GEMB01000486 JAS02641.1 GGFJ01010174 MBW59315.1 GBXI01003160 JAD11132.1 EZ419762 ACY69935.1 CH902635 EDV33817.1 GEZM01027480 JAV86723.1 CP012528 ALC48533.1 KB630508 KB631064 KB631065 ERL83640.1 ERL84055.1 ERL84057.1 GAMD01003307 JAA98283.1 GGFJ01010172 MBW59313.1 GGFM01001858 MBW22609.1 OUUW01000003 SPP77922.1 GAMC01001435 JAC05121.1 ADMH02000011 ETN68094.1 AK340198 BAH70851.1 ABLF02013658 CM000162 EDX02916.1 GFXV01007756 MBW19561.1 GGMR01011423 MBY24042.1
SOQ37222.1 AK402673 BAM19295.1 AK401355 BAM17977.1 AY829739 AAV91353.1 KZ149911 PZC78119.1 NWSH01001519 PCG70994.1 KQ459603 KPI92823.1 RSAL01000001 RVE55132.1 GAIX01014191 JAA78369.1 UFQS01001175 UFQS01001201 UFQT01001175 UFQT01001201 SSX09420.1 SSX09703.1 SSX29321.1 NNAY01002733 OXU20667.1 KQ971342 EFA04639.1 GL761674 EFZ22922.1 GALX01002995 JAB65471.1 KZ288310 PBC28610.1 JR048944 AEY60809.1 GDAI01001341 JAI16262.1 KQ976580 KYM79924.1 KQ434786 KZC05085.1 ADTU01014940 CVRI01000020 CRK91128.1 AXCN02000610 GECZ01001342 JAS68427.1 LBMM01002631 KMQ94565.1 GECZ01014164 JAS55605.1 GECU01011513 JAS96193.1 APCN01005209 AAAB01008964 EAA12214.5 APGK01016340 APGK01026055 APGK01026056 BT127480 KB740613 KB739824 AEE62442.1 ENN79996.1 ENN82108.1 KQ982335 KYQ57629.1 KQ794714 OAD47026.1 KK107750 QOIP01000012 EZA47909.1 RLU16007.1 KK107185 EZA55792.1 GFDL01007407 JAV27638.1 KQ435726 KOX78160.1 NEVH01020933 PNF20683.1 DS232187 EDS37024.1 GL449382 EFN82893.1 KQ981864 KYN34170.1 AAZX01007151 GDKW01000413 JAI56182.1 ACPB03021503 GAHY01001222 JAA76288.1 KQ979568 KYN20978.1 AJWK01003674 GFTR01002854 JAW13572.1 GECL01003249 JAP02875.1 GFDF01001791 JAV12293.1 KK852584 KDR20848.1 GL437917 EFN69882.1 AJVK01033225 PYGN01000224 PSN50752.1 GGMS01002205 MBY71408.1 GBBI01001201 JAC17511.1 GL888341 EGI62433.1 GGFK01003686 MBW37007.1 GEDC01022451 GEDC01002132 JAS14847.1 JAS35166.1 GEDC01029134 JAS08164.1 GEMB01000486 JAS02641.1 GGFJ01010174 MBW59315.1 GBXI01003160 JAD11132.1 EZ419762 ACY69935.1 CH902635 EDV33817.1 GEZM01027480 JAV86723.1 CP012528 ALC48533.1 KB630508 KB631064 KB631065 ERL83640.1 ERL84055.1 ERL84057.1 GAMD01003307 JAA98283.1 GGFJ01010172 MBW59313.1 GGFM01001858 MBW22609.1 OUUW01000003 SPP77922.1 GAMC01001435 JAC05121.1 ADMH02000011 ETN68094.1 AK340198 BAH70851.1 ABLF02013658 CM000162 EDX02916.1 GFXV01007756 MBW19561.1 GGMR01011423 MBY24042.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000283053
UP000215335
+ More
UP000007266 UP000005203 UP000242457 UP000078540 UP000075880 UP000076502 UP000075920 UP000005205 UP000183832 UP000075881 UP000075886 UP000036403 UP000075885 UP000075840 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000019118 UP000075809 UP000053097 UP000279307 UP000079169 UP000053105 UP000076408 UP000075901 UP000075900 UP000235965 UP000002320 UP000008237 UP000078541 UP000002358 UP000015103 UP000078492 UP000092461 UP000027135 UP000000311 UP000192223 UP000092462 UP000245037 UP000007755 UP000091820 UP000007801 UP000092553 UP000030742 UP000268350 UP000000673 UP000069272 UP000007819 UP000002282 UP000192221
UP000007266 UP000005203 UP000242457 UP000078540 UP000075880 UP000076502 UP000075920 UP000005205 UP000183832 UP000075881 UP000075886 UP000036403 UP000075885 UP000075840 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000019118 UP000075809 UP000053097 UP000279307 UP000079169 UP000053105 UP000076408 UP000075901 UP000075900 UP000235965 UP000002320 UP000008237 UP000078541 UP000002358 UP000015103 UP000078492 UP000092461 UP000027135 UP000000311 UP000192223 UP000092462 UP000245037 UP000007755 UP000091820 UP000007801 UP000092553 UP000030742 UP000268350 UP000000673 UP000069272 UP000007819 UP000002282 UP000192221
PRIDE
Pfam
PF02823 ATP-synt_DE_N
Interpro
Gene 3D
ProteinModelPortal
Q1HQ34
A0A194R1M3
A0A2H1VA11
I4DN05
I4DJ87
Q5MGP7
+ More
A0A2W1BST9 A0A2A4JHC9 A0A194PIE8 A0A3S2P226 S4NMX1 A0A336KXP7 A0A232EQS0 D6WNL2 E9I937 V5I9G1 A0A088AHV3 A0A2A3EA88 V9IKG8 A0A0K8TPE6 A0A151I243 A0A182JD00 A0A154NZJ1 A0A182W9Y8 A0A158NG24 A0A1J1HSS8 A0A182JTW4 A0A182QUP3 A0A1B6H198 A0A0J7NQ69 A0A1B6FZI8 A0A1B6JAZ1 A0A182P0H6 A0A182IEL1 A0A1S4GWD4 A0A182V6J7 A0A182UGZ6 A0A182WTI0 A0A182L2D6 Q7PNG6 J3JW62 A0A151XBC9 A0A310SBU4 A0A026VYH1 A0A026WIB6 A0A1Q3FJD4 A0A3Q0J2R9 A0A0M9A6D1 A0A182YKW5 A0A182SNK5 A0A1S3D4R1 A0A182R3B2 A0A2J7PWH3 B0WYE7 E2BMY9 A0A195F0R3 K7J447 A0A0P4VUG0 R4G8C3 A0A195E713 A0A1B0CAD3 A0A224XYC0 A0A0V0G4B2 A0A1L8E0M3 A0A067RCX9 E2A9J2 A0A1W4XKK8 A0A1B0EXQ7 A0A2P8Z2L9 A0A2S2Q0V8 A0A023F868 F4WTL2 A0A2M4A883 A0A1A9X3A7 A0A1B6EB44 A0A1B6C446 A0A161MDX5 A0A2M4C1Z6 A0A0A1XJK8 D1FPL2 B3MZL6 A0A1Y1MM15 A0A0M3QYZ4 U4U0R7 T1E770 A0A2M4C1Z3 A0A2M3Z256 A0A3B0JCU5 W8BUU2 W5JXC4 C4WSI1 A0A182FJU0 J9K2M7 B4PXG5 A0A2H8U0K5 A0A2S2P3X4 A0A1W4VV65
A0A2W1BST9 A0A2A4JHC9 A0A194PIE8 A0A3S2P226 S4NMX1 A0A336KXP7 A0A232EQS0 D6WNL2 E9I937 V5I9G1 A0A088AHV3 A0A2A3EA88 V9IKG8 A0A0K8TPE6 A0A151I243 A0A182JD00 A0A154NZJ1 A0A182W9Y8 A0A158NG24 A0A1J1HSS8 A0A182JTW4 A0A182QUP3 A0A1B6H198 A0A0J7NQ69 A0A1B6FZI8 A0A1B6JAZ1 A0A182P0H6 A0A182IEL1 A0A1S4GWD4 A0A182V6J7 A0A182UGZ6 A0A182WTI0 A0A182L2D6 Q7PNG6 J3JW62 A0A151XBC9 A0A310SBU4 A0A026VYH1 A0A026WIB6 A0A1Q3FJD4 A0A3Q0J2R9 A0A0M9A6D1 A0A182YKW5 A0A182SNK5 A0A1S3D4R1 A0A182R3B2 A0A2J7PWH3 B0WYE7 E2BMY9 A0A195F0R3 K7J447 A0A0P4VUG0 R4G8C3 A0A195E713 A0A1B0CAD3 A0A224XYC0 A0A0V0G4B2 A0A1L8E0M3 A0A067RCX9 E2A9J2 A0A1W4XKK8 A0A1B0EXQ7 A0A2P8Z2L9 A0A2S2Q0V8 A0A023F868 F4WTL2 A0A2M4A883 A0A1A9X3A7 A0A1B6EB44 A0A1B6C446 A0A161MDX5 A0A2M4C1Z6 A0A0A1XJK8 D1FPL2 B3MZL6 A0A1Y1MM15 A0A0M3QYZ4 U4U0R7 T1E770 A0A2M4C1Z3 A0A2M3Z256 A0A3B0JCU5 W8BUU2 W5JXC4 C4WSI1 A0A182FJU0 J9K2M7 B4PXG5 A0A2H8U0K5 A0A2S2P3X4 A0A1W4VV65
PDB
2W6J
E-value=6.15413e-25,
Score=277
Ontologies
PATHWAY
GO
PANTHER
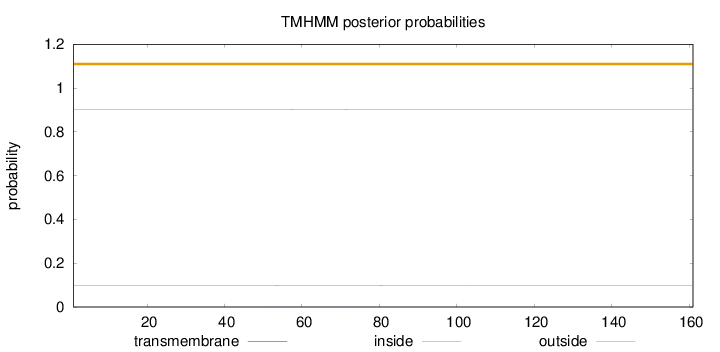
Topology
Length:
161
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00971
Exp number, first 60 AAs:
0.00291
Total prob of N-in:
0.09755
outside
1 - 161
Population Genetic Test Statistics
Pi
0.210938
Theta
2.269351
Tajima's D
-2.192294
CLR
1180.873863
CSRT
0.00169991500424979
Interpretation
Possibly Positive selection
