Pre Gene Modal
BGIBMGA010365
Annotation
uncharacterized_protein_LOC692518_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.16
Sequence
CDS
ATGGCGCCCAATGTAAAGGATGCAAATGGTGTTCTCTTTGAAAATGATGCTGATACTCCAGACTTAGGACTGTCCAGCACACCTGTACAGCAAGCCGACAACTACCCAAAGAAACTAGTTTGGAGGAATATAATACTTTTTACATACCTTCATATTGCTGCGTTGTATGGAGGCTACCTTTTCTTATTTCATGCGAAATGGCAAACAGACCTATTTGCCTATATATTGTATGTAATGTCAGGACTTGGCATCACAGCAGGTGCACATAGATTATGGGCACATAAATCATACAAAGCAAAGTGGCCTCTACGTGTTATCCTTATCATCTTCAATTCCTTGGCTTTTCAGGATTCTGCTTTAGATTGGGCACGTGATCATCGCATGCATCATAAATACTCCGAGACTGATGCCGACCCACATAATGCCACACGAGGTTTCTTTTTCTCTCACATTGGCTGGCTTCTAGTGAGGAAACATCCAGAACTGAAGAGAAAGGGCAAGGGTCTCGACCTAAGTGACTTATATGCTGATCCCATTCTTCGATTCCAGAAAAAGTACTATTTAATTCTGATGCCAATCGCTTGCTTCATTTTGCCAACAGTGATTCCGGTGTATTTTTGGAATGAGTCTTGGATTAACGCGTTCTTCGTTGCGACTCTTTTCCGTTATACATTTATTTTGAATGTCACATGGCTAGTGAACTCCGCTGCTCACAAATGGGGTGGAAAACCCTACGACAAGAATATCAAGCCATCCGAGAACATCTCCGTGTCGGTGTTTGCACTCGGCGAGGGATTCCACAACTACCACCACACATTCCCATGGGACTACAAGACAGCGGAACTCGGTAACAACAGATTAAACTTCACCACCAATTTCATCAATTTCTTTGCTAAAATTGGATGGGCATACGATTTGAAAACTGTATCTGACGAGATCATAAAGAACCGTGTGAACCGCACCGGTGATGGCTCTCACTACCTCTGGGGCTGGGGCGATAAAGATCTGGACAAAGAGGAAATAAAGCAAGCCATCCGCATCAATCCTAAGGACAATTGA
Protein
MAPNVKDANGVLFENDADTPDLGLSSTPVQQADNYPKKLVWRNIILFTYLHIAALYGGYLFLFHAKWQTDLFAYILYVMSGLGITAGAHRLWAHKSYKAKWPLRVILIIFNSLAFQDSALDWARDHRMHHKYSETDADPHNATRGFFFSHIGWLLVRKHPELKRKGKGLDLSDLYADPILRFQKKYYLILMPIACFILPTVIPVYFWNESWINAFFVATLFRYTFILNVTWLVNSAAHKWGGKPYDKNIKPSENISVSVFALGEGFHNYHHTFPWDYKTAELGNNRLNFTTNFINFFAKIGWAYDLKTVSDEIIKNRVNRTGDGSHYLWGWGDKDLDKEEIKQAIRINPKDN
Summary
Cofactor
Fe cation
Similarity
Belongs to the fatty acid desaturase type 1 family.
Uniprot
I3QPX9
Q75PL6
H9JLG4
Q4A182
S4PEM6
A0A212FG51
+ More
A0A194R0B7 A0A194PHD4 B7SB75 Q7K403 Q9NGQ7 B7SB77 B7SB76 A0A0M3L9S4 A0A1E1VYJ4 D8UUT5 Q9NB25 A0A0U2F3B9 A0A291P0G3 Q8WQQ0 Q8ITE5 Q8ITE4 T1RTF5 Q8ITE8 A0A0U2F3B3 Q8MZZ4 T1RTD3 E3TMV0 A0A2H1V8Q0 A0A1L8D6B6 U5KFT8 A9YRX5 I7B370 A0A0P0EGH8 Q9NGQ8 A9ZSZ7 G8FQ63 A0A076FR53 G8FQ64 Q6US82 Q9NG28 G8FQ62 B6CBS2 A0A0L7LB21 A0A1A9Z238 D3TP66 A0A1A9WBD6 A0A182QY62 A0A182MQV9 A0A182PSJ2 A0A182Y3S6 A0A2M3YY30 A0A182VZL7 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 W5J215 A0A182NRU1 B4QSA3 B3M2R7 A0A182RZM3 A0A2M3YY13 Q9U972 Q7K4Y0 Q94541 A0A182K5W6 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 B4PPA5 T1DNF2 Q9U967 B4GZ70 A0A182IX38 A6P6Z1 Q29B20 A0A1I8NWA7 A0A023EPC8 A0A0K8TSL7 Q17G31 Q8ITN6 T1PCA2 W6ACM3 B3P4B3 A0A1L8EEG9 B0X4F1 A0A084W6J7 B4JRQ4 B4M0C1 A0A1A9WBD7 B4K744 T1PL97 A0A0M4EPW0 T1E253 A0A1Q3FJ73 A0A1E1WUE8 A0A1B0CEY2 A0A336M2F9
A0A194R0B7 A0A194PHD4 B7SB75 Q7K403 Q9NGQ7 B7SB77 B7SB76 A0A0M3L9S4 A0A1E1VYJ4 D8UUT5 Q9NB25 A0A0U2F3B9 A0A291P0G3 Q8WQQ0 Q8ITE5 Q8ITE4 T1RTF5 Q8ITE8 A0A0U2F3B3 Q8MZZ4 T1RTD3 E3TMV0 A0A2H1V8Q0 A0A1L8D6B6 U5KFT8 A9YRX5 I7B370 A0A0P0EGH8 Q9NGQ8 A9ZSZ7 G8FQ63 A0A076FR53 G8FQ64 Q6US82 Q9NG28 G8FQ62 B6CBS2 A0A0L7LB21 A0A1A9Z238 D3TP66 A0A1A9WBD6 A0A182QY62 A0A182MQV9 A0A182PSJ2 A0A182Y3S6 A0A2M3YY30 A0A182VZL7 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 W5J215 A0A182NRU1 B4QSA3 B3M2R7 A0A182RZM3 A0A2M3YY13 Q9U972 Q7K4Y0 Q94541 A0A182K5W6 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 B4PPA5 T1DNF2 Q9U967 B4GZ70 A0A182IX38 A6P6Z1 Q29B20 A0A1I8NWA7 A0A023EPC8 A0A0K8TSL7 Q17G31 Q8ITN6 T1PCA2 W6ACM3 B3P4B3 A0A1L8EEG9 B0X4F1 A0A084W6J7 B4JRQ4 B4M0C1 A0A1A9WBD7 B4K744 T1PL97 A0A0M4EPW0 T1E253 A0A1Q3FJ73 A0A1E1WUE8 A0A1B0CEY2 A0A336M2F9
Pubmed
19121390
23622113
22118469
26354079
20403437
12237399
+ More
11483431 28986331 28756777 11805319 15527989 24500170 15455060 12524345 12770579 20691782 24053512 18405835 26445454 24211206 22291612 24817326 15544945 12213232 18831750 26227816 20353571 25244985 20920257 23761445 17994087 18057021 10920187 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9501419 20966253 12364791 14747013 17210077 17550304 17916502 15632085 23185243 24945155 26369729 17510324 12421411 25315136 24438588 24330624
11483431 28986331 28756777 11805319 15527989 24500170 15455060 12524345 12770579 20691782 24053512 18405835 26445454 24211206 22291612 24817326 15544945 12213232 18831750 26227816 20353571 25244985 20920257 23761445 17994087 18057021 10920187 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9501419 20966253 12364791 14747013 17210077 17550304 17916502 15632085 23185243 24945155 26369729 17510324 12421411 25315136 24438588 24330624
EMBL
JQ729998
AFK13830.1
JQ729997
AB166852
AFK13829.1
BAD18123.1
+ More
BABH01005826 AM076338 CAJ27975.1 GAIX01006995 JAA85565.1 AGBW02008720 OWR52703.1 KQ460883 KPJ11142.1 KQ459603 KPI92821.1 EU152401 ABX71810.1 AY057862 AAL27033.1 AF243047 AAF44710.1 EU152403 ABX71812.1 EU152402 ABX71811.1 KM366791 AIM40221.1 GDQN01011259 GDQN01009722 GDQN01007857 JAT79795.1 JAT81332.1 JAT83197.1 GQ225747 GQ225749 ADE58520.1 AF272343 AAF81788.1 KP008120 MF687582 AKU76407.1 ATJ44508.1 MF687527 KZ149911 ATJ44453.1 PZC78117.1 AY061988 AAL35750.1 AF518020 AAN39700.1 AF518021 AAN39701.1 JX531653 NWSH01001519 AGO45838.1 PCG70996.1 AF518017 AAN39697.1 KP008113 AKU76400.1 AF482906 AAM28481.2 JX531660 AGO45845.1 GU952765 ADO85598.1 ODYU01001259 SOQ37220.1 GEYN01000137 JAV01992.1 JX989152 AGR49311.1 EU285579 ABX90048.1 JX126483 AFO38464.1 KT261693 ALJ30233.1 AF243046 AAF44709.1 AB373031 BAF97042.1 JN022473 AER29842.1 KF960740 AII21942.1 JN022474 JN022475 AER29843.1 AY362878 AAQ74258.1 AF268275 JN022476 AAF73073.1 AER29845.1 JN022472 AER29841.1 EU152332 ABX71627.1 JTDY01001896 KOB72605.1 CCAG010012100 EZ423218 ADD19494.1 AXCN02001148 AXCM01000026 GGFM01000428 MBW21179.1 GGFK01006347 MBW39668.1 GGFJ01006787 MBW55928.1 ADMH02002143 ETN58292.1 CM000364 EDX13199.1 CH902617 EDV42388.1 KPU79686.1 KPU79687.1 GGFM01000421 MBW21172.1 AJ245747 CAB52474.1 AE014297 AY051593 AAF54819.1 AAF54820.1 AAF54821.1 AAF54822.1 AAK93017.1 AAN13544.1 U73160 AAB17283.1 AAAB01008987 EAA43119.4 EGK97500.1 EGK97501.1 EGK97502.1 APCN01000909 CM000160 EDW97111.1 KRK03534.1 GAMD01002940 JAA98650.1 AJ245748 CAB52475.1 CH479198 EDW28088.1 AB300221 BAF69118.1 CM000070 EAL27178.1 KRS99817.1 GAPW01002266 JAC11332.1 GDAI01000236 JAI17367.1 CH477266 EAT45554.1 EAT45555.1 AF417841 AAN31393.1 KA646397 AFP61026.1 KF835695 AHI54575.1 CH954181 EDV49358.1 GFDG01001755 JAV17044.1 DS232338 EDS40304.1 ATLV01020821 KE525308 KFB45841.1 CH916373 EDV94444.1 CH940650 EDW67283.1 KRF83228.1 CH933806 EDW16357.1 KRG02060.1 KRG02061.1 KA649511 KA649512 AFP64140.1 CP012526 ALC47890.1 GALA01001044 JAA93808.1 GFDL01007467 JAV27578.1 GDQN01000462 JAT90592.1 AJWK01009463 UFQT01000109 SSX20158.1
BABH01005826 AM076338 CAJ27975.1 GAIX01006995 JAA85565.1 AGBW02008720 OWR52703.1 KQ460883 KPJ11142.1 KQ459603 KPI92821.1 EU152401 ABX71810.1 AY057862 AAL27033.1 AF243047 AAF44710.1 EU152403 ABX71812.1 EU152402 ABX71811.1 KM366791 AIM40221.1 GDQN01011259 GDQN01009722 GDQN01007857 JAT79795.1 JAT81332.1 JAT83197.1 GQ225747 GQ225749 ADE58520.1 AF272343 AAF81788.1 KP008120 MF687582 AKU76407.1 ATJ44508.1 MF687527 KZ149911 ATJ44453.1 PZC78117.1 AY061988 AAL35750.1 AF518020 AAN39700.1 AF518021 AAN39701.1 JX531653 NWSH01001519 AGO45838.1 PCG70996.1 AF518017 AAN39697.1 KP008113 AKU76400.1 AF482906 AAM28481.2 JX531660 AGO45845.1 GU952765 ADO85598.1 ODYU01001259 SOQ37220.1 GEYN01000137 JAV01992.1 JX989152 AGR49311.1 EU285579 ABX90048.1 JX126483 AFO38464.1 KT261693 ALJ30233.1 AF243046 AAF44709.1 AB373031 BAF97042.1 JN022473 AER29842.1 KF960740 AII21942.1 JN022474 JN022475 AER29843.1 AY362878 AAQ74258.1 AF268275 JN022476 AAF73073.1 AER29845.1 JN022472 AER29841.1 EU152332 ABX71627.1 JTDY01001896 KOB72605.1 CCAG010012100 EZ423218 ADD19494.1 AXCN02001148 AXCM01000026 GGFM01000428 MBW21179.1 GGFK01006347 MBW39668.1 GGFJ01006787 MBW55928.1 ADMH02002143 ETN58292.1 CM000364 EDX13199.1 CH902617 EDV42388.1 KPU79686.1 KPU79687.1 GGFM01000421 MBW21172.1 AJ245747 CAB52474.1 AE014297 AY051593 AAF54819.1 AAF54820.1 AAF54821.1 AAF54822.1 AAK93017.1 AAN13544.1 U73160 AAB17283.1 AAAB01008987 EAA43119.4 EGK97500.1 EGK97501.1 EGK97502.1 APCN01000909 CM000160 EDW97111.1 KRK03534.1 GAMD01002940 JAA98650.1 AJ245748 CAB52475.1 CH479198 EDW28088.1 AB300221 BAF69118.1 CM000070 EAL27178.1 KRS99817.1 GAPW01002266 JAC11332.1 GDAI01000236 JAI17367.1 CH477266 EAT45554.1 EAT45555.1 AF417841 AAN31393.1 KA646397 AFP61026.1 KF835695 AHI54575.1 CH954181 EDV49358.1 GFDG01001755 JAV17044.1 DS232338 EDS40304.1 ATLV01020821 KE525308 KFB45841.1 CH916373 EDV94444.1 CH940650 EDW67283.1 KRF83228.1 CH933806 EDW16357.1 KRG02060.1 KRG02061.1 KA649511 KA649512 AFP64140.1 CP012526 ALC47890.1 GALA01001044 JAA93808.1 GFDL01007467 JAV27578.1 GDQN01000462 JAT90592.1 AJWK01009463 UFQT01000109 SSX20158.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000037510
+ More
UP000092445 UP000092444 UP000091820 UP000075886 UP000075883 UP000075885 UP000076408 UP000075920 UP000069272 UP000000673 UP000075884 UP000000304 UP000007801 UP000075900 UP000000803 UP000075881 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000002282 UP000008744 UP000075880 UP000001819 UP000095300 UP000008820 UP000095301 UP000008711 UP000002320 UP000030765 UP000001070 UP000008792 UP000009192 UP000092553 UP000092461
UP000092445 UP000092444 UP000091820 UP000075886 UP000075883 UP000075885 UP000076408 UP000075920 UP000069272 UP000000673 UP000075884 UP000000304 UP000007801 UP000075900 UP000000803 UP000075881 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000002282 UP000008744 UP000075880 UP000001819 UP000095300 UP000008820 UP000095301 UP000008711 UP000002320 UP000030765 UP000001070 UP000008792 UP000009192 UP000092553 UP000092461
Pfam
PF00487 FA_desaturase
Interpro
ProteinModelPortal
I3QPX9
Q75PL6
H9JLG4
Q4A182
S4PEM6
A0A212FG51
+ More
A0A194R0B7 A0A194PHD4 B7SB75 Q7K403 Q9NGQ7 B7SB77 B7SB76 A0A0M3L9S4 A0A1E1VYJ4 D8UUT5 Q9NB25 A0A0U2F3B9 A0A291P0G3 Q8WQQ0 Q8ITE5 Q8ITE4 T1RTF5 Q8ITE8 A0A0U2F3B3 Q8MZZ4 T1RTD3 E3TMV0 A0A2H1V8Q0 A0A1L8D6B6 U5KFT8 A9YRX5 I7B370 A0A0P0EGH8 Q9NGQ8 A9ZSZ7 G8FQ63 A0A076FR53 G8FQ64 Q6US82 Q9NG28 G8FQ62 B6CBS2 A0A0L7LB21 A0A1A9Z238 D3TP66 A0A1A9WBD6 A0A182QY62 A0A182MQV9 A0A182PSJ2 A0A182Y3S6 A0A2M3YY30 A0A182VZL7 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 W5J215 A0A182NRU1 B4QSA3 B3M2R7 A0A182RZM3 A0A2M3YY13 Q9U972 Q7K4Y0 Q94541 A0A182K5W6 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 B4PPA5 T1DNF2 Q9U967 B4GZ70 A0A182IX38 A6P6Z1 Q29B20 A0A1I8NWA7 A0A023EPC8 A0A0K8TSL7 Q17G31 Q8ITN6 T1PCA2 W6ACM3 B3P4B3 A0A1L8EEG9 B0X4F1 A0A084W6J7 B4JRQ4 B4M0C1 A0A1A9WBD7 B4K744 T1PL97 A0A0M4EPW0 T1E253 A0A1Q3FJ73 A0A1E1WUE8 A0A1B0CEY2 A0A336M2F9
A0A194R0B7 A0A194PHD4 B7SB75 Q7K403 Q9NGQ7 B7SB77 B7SB76 A0A0M3L9S4 A0A1E1VYJ4 D8UUT5 Q9NB25 A0A0U2F3B9 A0A291P0G3 Q8WQQ0 Q8ITE5 Q8ITE4 T1RTF5 Q8ITE8 A0A0U2F3B3 Q8MZZ4 T1RTD3 E3TMV0 A0A2H1V8Q0 A0A1L8D6B6 U5KFT8 A9YRX5 I7B370 A0A0P0EGH8 Q9NGQ8 A9ZSZ7 G8FQ63 A0A076FR53 G8FQ64 Q6US82 Q9NG28 G8FQ62 B6CBS2 A0A0L7LB21 A0A1A9Z238 D3TP66 A0A1A9WBD6 A0A182QY62 A0A182MQV9 A0A182PSJ2 A0A182Y3S6 A0A2M3YY30 A0A182VZL7 A0A2M4AGB5 A0A182FAQ2 A0A2M4BT31 W5J215 A0A182NRU1 B4QSA3 B3M2R7 A0A182RZM3 A0A2M3YY13 Q9U972 Q7K4Y0 Q94541 A0A182K5W6 A0A182UTT3 A0A182WSS9 A0A182LA27 F5HMH9 A0A182HLN2 B4PPA5 T1DNF2 Q9U967 B4GZ70 A0A182IX38 A6P6Z1 Q29B20 A0A1I8NWA7 A0A023EPC8 A0A0K8TSL7 Q17G31 Q8ITN6 T1PCA2 W6ACM3 B3P4B3 A0A1L8EEG9 B0X4F1 A0A084W6J7 B4JRQ4 B4M0C1 A0A1A9WBD7 B4K744 T1PL97 A0A0M4EPW0 T1E253 A0A1Q3FJ73 A0A1E1WUE8 A0A1B0CEY2 A0A336M2F9
PDB
4YMK
E-value=6.29955e-97,
Score=903
Ontologies
PATHWAY
GO
GO:0016717
GO:0016021
GO:0006633
GO:0004768
GO:0042593
GO:0040018
GO:0005783
GO:0019216
GO:0048047
GO:0006636
GO:0031410
GO:0045793
GO:0010506
GO:0006723
GO:0042594
GO:0045572
GO:0009744
GO:0070328
GO:0060179
GO:0042811
GO:0005506
GO:0005789
GO:0006629
GO:0003676
GO:0008270
GO:0006520
GO:0006508
GO:0008237
GO:0043401
GO:0003707
GO:0051537
GO:0055114
PANTHER
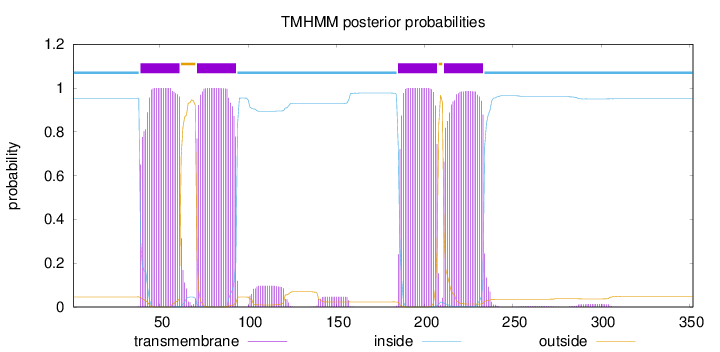
Topology
Length:
352
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
91.5661600000001
Exp number, first 60 AAs:
20.78937
Total prob of N-in:
0.95393
POSSIBLE N-term signal
sequence
inside
1 - 38
TMhelix
39 - 61
outside
62 - 70
TMhelix
71 - 93
inside
94 - 184
TMhelix
185 - 207
outside
208 - 210
TMhelix
211 - 233
inside
234 - 352
Population Genetic Test Statistics
Pi
0.229116
Theta
1.472672
Tajima's D
-1.823006
CLR
439.390715
CSRT
0.0167991600419979
Interpretation
Possibly Positive selection
