Gene
KWMTBOMO07244 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010586
Annotation
hypothetical_protein_KGM_20562_[Danaus_plexippus]
Full name
Palmitoyltransferase
Location in the cell
PlasmaMembrane Reliability : 2.808
Sequence
CDS
ATGTCGGAATCGTGTCAGGACGGTAATTGTCAAGGAAATAATGAATATGGACCTGAAGAATCGCCTACCAAACTCATTGTCAATTACATTCCTGAAGTCATGACCCAGGATATGATGTTTTCTTTATTTTCAACAATGGGCAAATTGGAGAGTTGTAAGTTGATAGCAAATCGAGGCTATGGATTCGTCGAGTACACACGTCCGGACGATGCAGTTAAAGCTCGTAAAGCGTTCAACGGTCTACTGATGCAGAATAAGACTCTCAAAGTGTCACACGCATTACTTAATCCAGAACTTAAGCCACCAACAAAACCTGAAGCCGACTGGAATCTCTACATTTGTAACCTACCTAATGAATTGACCTTGCAAGTGAGGCCTTATCAATGGATTAATCATATTAGTGCTGTGGGTGAACATAATTCACCAAATACATGGTCTATTTATATTTATAATATTGCTTTGGAAGTAGAAGAGTTAACTCTTTGGCAACTTTTCGGTCCATATGGTGCTATTGTTTCAGTTAAGATTATTAGGGATCATCAGACTAATAAAAGTAAAGGATATGGTTTTGTTACAATGAGAAATTACGACCAAGCTGCGATGGCAATACAAGCATTAAACGGATATCTATTGCACGGTCAACCTCTCTCTATTTTCGGTAAAGAGGCGCTGGTCGTGGCGGACTGCGAGAACTCTGCTGTAGCGGGGGGCGTCGGCCTCGCCGTGAGTAGCAAGGTATATCCAAGTCCGGGGGCGGCGCGCGGCGTGTGGGCGCTGCCACAGCCGCCCCCGTGCACGATGGCCGCAGAGGAATCTCTCCCACAGGGCTCACCCATGTCGCCGGATGCTCCAGTCGCACCACCCAGGACTATCACGAGTTTCAATCAATACTCCGATTTGCACCAACTAATATTTGAGGCGGTTCGATCAGGAGAAGTATCGGAAATCGAACGCCTGGTTGAAAAACTTGGAGTGGAAGTCTTGAATGCTCGGGACCAGCATGGCTATACTCCCGCACACTGGGCATCGCTTGATGGGAGCGTGGCGGTCATGCGATACCTCGTGGAAAGATCAGCTCCTGTCGATCTTTCCTGTCTTGGCACTCAGGGGCCGCGTCCGATACACTGGGCTTGCCGCAAAGGACATGCATCTATCGTTCAAGTTCTCCTTCAGGCCGGTGTTGCTGTCAATGCTGCAGATTTCAAAGGTTTGACACCTTTAATGACAGCTTGCATGTATGGTAAAACAGCCACAGCTGCTTATCTGTTAGGAATGGGTGCAGCTACAAGACTGTCTGATATAAACGGAGACACAGCCCTACATTGGGCGGCATATAAGGGACATGCAGATGTCGTTCGTCTTTTAATTTACTCTGGAGTTCCATTACACTGTACTGATAATTTTGGTTCGACACCCCTACATTTAGCATGTTTGTCGGGAAACTTAACTTGTGTTAGATTACTCTGTGAAAAGGTAAAGGCTGAGTTAGAACCGCGCGATAAAAATGGAAAAACACCGCTAATGCTAGCACAGAGCCACAGGCATGCAGAAGTTGTGAAATTACTGCAAAAGGAAATGAAACGTAAATCCCATTGGATACCCCCACTGTCAGAATTATGGGCGTTATTGTTCGGCGGCGCTGGTGATTCTAAAGGGCCACTGTTGTTCTTTTTAATCTCAGTTTTATTGTGGGGTTATCCTATACGTGATCCCGGATACGTGCCGCAGAATTCGGAAACATACTACAGAGCTATAAGACAAATCCCTTATTACGATAAATGGAAGAAGAGAAATATTATATTATCAAGACTGTGTCATACGTGTCGATGCTTAAGACCTCTGAGAGCAAAACATTGTCGCATCTGCAAACGTTGCGTAGCGTATTTTGACCATCACTGCCCGTTTATCTACAACTGTGTCGGAGTTCGCAATCGAATGTGGTTCTTCTTGTTCGTGATGAGCGTTGCGATCAACTGCACATTGTCGATCTACTTTGCCTGTTATTGTCTACTACTCGAGGGATTCGGTCTGCTGTATATGCTGGGGCTACTTGAAGCTATTACTTTTTGTGCCCTCGGTTGGATTTTGACTTGCACTTCGGTGCTGCACGCCTGTATGAACTTAACCACAAACGAAATGTTTAATTACAAACGATATCCATACTTGAGGGACAAACGAGGAAGATACCAAAATCCATTTTCACGTGGACCCATCATGAATCTGTTCGAGTTTTTCGTGTGCCTGCCGGATAAATGCGACGACCATGACATTTTTCACGAAGAAAGTATATAA
Protein
MSESCQDGNCQGNNEYGPEESPTKLIVNYIPEVMTQDMMFSLFSTMGKLESCKLIANRGYGFVEYTRPDDAVKARKAFNGLLMQNKTLKVSHALLNPELKPPTKPEADWNLYICNLPNELTLQVRPYQWINHISAVGEHNSPNTWSIYIYNIALEVEELTLWQLFGPYGAIVSVKIIRDHQTNKSKGYGFVTMRNYDQAAMAIQALNGYLLHGQPLSIFGKEALVVADCENSAVAGGVGLAVSSKVYPSPGAARGVWALPQPPPCTMAAEESLPQGSPMSPDAPVAPPRTITSFNQYSDLHQLIFEAVRSGEVSEIERLVEKLGVEVLNARDQHGYTPAHWASLDGSVAVMRYLVERSAPVDLSCLGTQGPRPIHWACRKGHASIVQVLLQAGVAVNAADFKGLTPLMTACMYGKTATAAYLLGMGAATRLSDINGDTALHWAAYKGHADVVRLLIYSGVPLHCTDNFGSTPLHLACLSGNLTCVRLLCEKVKAELEPRDKNGKTPLMLAQSHRHAEVVKLLQKEMKRKSHWIPPLSELWALLFGGAGDSKGPLLFFLISVLLWGYPIRDPGYVPQNSETYYRAIRQIPYYDKWKKRNIILSRLCHTCRCLRPLRAKHCRICKRCVAYFDHHCPFIYNCVGVRNRMWFFLFVMSVAINCTLSIYFACYCLLLEGFGLLYMLGLLEAITFCALGWILTCTSVLHACMNLTTNEMFNYKRYPYLRDKRGRYQNPFSRGPIMNLFEFFVCLPDKCDDHDIFHEESI
Summary
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Similarity
Belongs to the DHHC palmitoyltransferase family.
Uniprot
A0A2H1V8T3
A0A212FG42
H9JM34
A0A2A4JFZ7
A0A2W1BZH8
A0A194R019
+ More
A0A194PNW7 A0A0L7LB15 A0A084VL20 A0A182PDV8 A0A182XNS6 A0A182F647 A0A182JEN3 A0A182R155 Q7PWK8 Q17LW6 A0A182UUW1 A0A182LMR8 B0WB14 B4LSF8 B5DHZ9 B3MVH4 A0A1W4VBZ7 A0A0J9R186 B4P1Y8 B3N3X0 Q9VKB1 B4IE66 B4N775 A0A0M4E5I5 A0A0L0CMQ1 A0A1A9Z7X8 A0A195AYF8 A0A0L7RGX8 E9JD72 T1I5S7 A0A2A3EDW4 A0A0N0BF79 E2CAD7 A0A232F461 A0A336M051 A0A310SKS7 B4KI04 A0A087ZYY1
A0A194PNW7 A0A0L7LB15 A0A084VL20 A0A182PDV8 A0A182XNS6 A0A182F647 A0A182JEN3 A0A182R155 Q7PWK8 Q17LW6 A0A182UUW1 A0A182LMR8 B0WB14 B4LSF8 B5DHZ9 B3MVH4 A0A1W4VBZ7 A0A0J9R186 B4P1Y8 B3N3X0 Q9VKB1 B4IE66 B4N775 A0A0M4E5I5 A0A0L0CMQ1 A0A1A9Z7X8 A0A195AYF8 A0A0L7RGX8 E9JD72 T1I5S7 A0A2A3EDW4 A0A0N0BF79 E2CAD7 A0A232F461 A0A336M051 A0A310SKS7 B4KI04 A0A087ZYY1
EC Number
2.3.1.225
Pubmed
EMBL
ODYU01001259
SOQ37219.1
AGBW02008720
OWR52704.1
BABH01005825
BABH01005826
+ More
NWSH01001519 PCG70997.1 KZ149911 PZC78116.1 KQ460883 KPJ11143.1 KQ459603 KPI92820.1 JTDY01001896 KOB72600.1 ATLV01014396 ATLV01014397 KE524970 KFB38664.1 AXCN02000795 AAAB01008984 EAA14821.2 CH477210 EAT47705.1 DS231876 EDS41972.1 CH940649 EDW64780.2 CH379060 EDY69936.2 CH902624 EDV33239.2 CM002910 KMY89878.1 CM000157 EDW88159.1 CH954177 EDV58822.1 AE014134 AY058389 AAF53165.1 AAL13618.1 CH480831 EDW45893.1 CH964182 EDW80216.2 CP012523 ALC39384.1 JRES01000186 KNC33492.1 KQ976701 KYM77231.1 KQ414596 KOC70089.1 GL771866 EFZ09193.1 ACPB03003574 KZ288269 PBC29973.1 KQ435809 KOX72968.1 GL453975 EFN75103.1 NNAY01001055 OXU25319.1 UFQS01000238 UFQT01000238 SSX01836.1 SSX22213.1 KQ760243 OAD61281.1 CH933807 EDW11286.2
NWSH01001519 PCG70997.1 KZ149911 PZC78116.1 KQ460883 KPJ11143.1 KQ459603 KPI92820.1 JTDY01001896 KOB72600.1 ATLV01014396 ATLV01014397 KE524970 KFB38664.1 AXCN02000795 AAAB01008984 EAA14821.2 CH477210 EAT47705.1 DS231876 EDS41972.1 CH940649 EDW64780.2 CH379060 EDY69936.2 CH902624 EDV33239.2 CM002910 KMY89878.1 CM000157 EDW88159.1 CH954177 EDV58822.1 AE014134 AY058389 AAF53165.1 AAL13618.1 CH480831 EDW45893.1 CH964182 EDW80216.2 CP012523 ALC39384.1 JRES01000186 KNC33492.1 KQ976701 KYM77231.1 KQ414596 KOC70089.1 GL771866 EFZ09193.1 ACPB03003574 KZ288269 PBC29973.1 KQ435809 KOX72968.1 GL453975 EFN75103.1 NNAY01001055 OXU25319.1 UFQS01000238 UFQT01000238 SSX01836.1 SSX22213.1 KQ760243 OAD61281.1 CH933807 EDW11286.2
Proteomes
UP000007151
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
+ More
UP000030765 UP000075885 UP000076407 UP000069272 UP000075880 UP000075886 UP000007062 UP000008820 UP000075903 UP000075882 UP000002320 UP000008792 UP000001819 UP000007801 UP000192221 UP000002282 UP000008711 UP000000803 UP000001292 UP000007798 UP000092553 UP000037069 UP000092445 UP000078540 UP000053825 UP000015103 UP000242457 UP000053105 UP000008237 UP000215335 UP000009192 UP000005203
UP000030765 UP000075885 UP000076407 UP000069272 UP000075880 UP000075886 UP000007062 UP000008820 UP000075903 UP000075882 UP000002320 UP000008792 UP000001819 UP000007801 UP000192221 UP000002282 UP000008711 UP000000803 UP000001292 UP000007798 UP000092553 UP000037069 UP000092445 UP000078540 UP000053825 UP000015103 UP000242457 UP000053105 UP000008237 UP000215335 UP000009192 UP000005203
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1V8T3
A0A212FG42
H9JM34
A0A2A4JFZ7
A0A2W1BZH8
A0A194R019
+ More
A0A194PNW7 A0A0L7LB15 A0A084VL20 A0A182PDV8 A0A182XNS6 A0A182F647 A0A182JEN3 A0A182R155 Q7PWK8 Q17LW6 A0A182UUW1 A0A182LMR8 B0WB14 B4LSF8 B5DHZ9 B3MVH4 A0A1W4VBZ7 A0A0J9R186 B4P1Y8 B3N3X0 Q9VKB1 B4IE66 B4N775 A0A0M4E5I5 A0A0L0CMQ1 A0A1A9Z7X8 A0A195AYF8 A0A0L7RGX8 E9JD72 T1I5S7 A0A2A3EDW4 A0A0N0BF79 E2CAD7 A0A232F461 A0A336M051 A0A310SKS7 B4KI04 A0A087ZYY1
A0A194PNW7 A0A0L7LB15 A0A084VL20 A0A182PDV8 A0A182XNS6 A0A182F647 A0A182JEN3 A0A182R155 Q7PWK8 Q17LW6 A0A182UUW1 A0A182LMR8 B0WB14 B4LSF8 B5DHZ9 B3MVH4 A0A1W4VBZ7 A0A0J9R186 B4P1Y8 B3N3X0 Q9VKB1 B4IE66 B4N775 A0A0M4E5I5 A0A0L0CMQ1 A0A1A9Z7X8 A0A195AYF8 A0A0L7RGX8 E9JD72 T1I5S7 A0A2A3EDW4 A0A0N0BF79 E2CAD7 A0A232F461 A0A336M051 A0A310SKS7 B4KI04 A0A087ZYY1
PDB
6MOK
E-value=2.72527e-32,
Score=349
Ontologies
GO
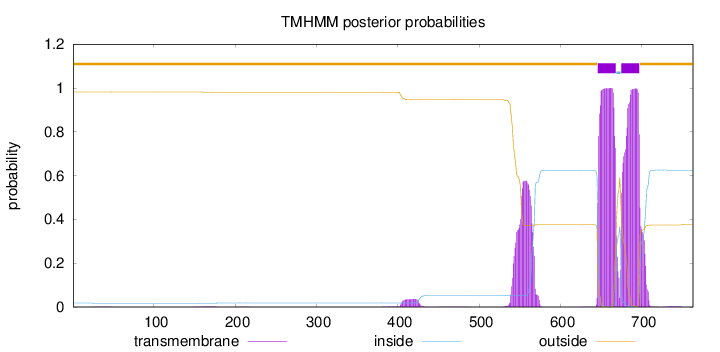
Topology
Length:
763
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
58.54982
Exp number, first 60 AAs:
0.00562
Total prob of N-in:
0.01781
outside
1 - 645
TMhelix
646 - 668
inside
669 - 674
TMhelix
675 - 697
outside
698 - 763
Population Genetic Test Statistics
Pi
2.776778
Theta
3.357768
Tajima's D
-0.950028
CLR
4.704824
CSRT
0.146042697865107
Interpretation
Uncertain
