Gene
KWMTBOMO07243
Pre Gene Modal
BGIBMGA010585
Annotation
hypothetical_protein_KGM_20561_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.95
Sequence
CDS
ATGAGCGATGAAAAACAGCCACTCCTAACTGGACCAAACAGTCCAGAAAATGTAATTGAAATACAATCAACCGAACCAGTGATAAATGATGATGGTCCTTCAACTGAGAAAACAAAAGGCGACTATTGCCCTGCATCAGAAAGGTGTTTAGAATATCCAACATCAAACTTTGATACAATGATTCATCTTCTTAAAGGTAATATAGGAACGGGGATACTAGCTATGCCTGACGCCTTCAAAAATGCGGGACTTATTTTTGGTGTATTTTGCACATTGATTATGGGTGCTATGTGCACTCATTGCATGCACATATTAGTACAGTGTTCTCATGAACTATGCATAAGAAGTGAAAAACCAGCTTTGAGCTTCTCCGAAGTTATTGAGGACTCGTTTTTGTCAGGACCGATTGTATTTCGACCATATGCAAAGAAAATGAGGGCATTAGTCAATATATTTCTCGTTATAACGCAACTGGGATTTTGCTGTGTGTATTTTTTATTTGTCGCTACTAACCTTCAAGATACAATGCATTTATTTCGTATAAATCTAAGCGTACATTTGTACCTGACTCTGTTATTTCCGCTAATCGTCGCATTAGCTATGGTAAAAAATTTAAAATATTTAACACCAGTGTCTCTGGTGGCATCCATAATGACAGCTTGGGGCCTAGCTATCACGTTCTACTATATTCTTCAAGATTTGCCGCACTCTAAAGAAGTGAACCCGATAGCCACATGGCACCAGTTACCATTGTATTTCGGTACAGCAATATATGCGTTCGAAGGAATTGGAGTGGTATTACCTCTGGAAAATAACATGAAAACTCCTGAAGACTTTGGTGGATGGAACGGCGTTCTGAACACGGGTATGGTCATAGTGGCAGCTCTGTACACCGCAGTTGGATTCTTTGGTTATTTAAAATACGGAGACAGCGTCCAAGGTAGCATAACTCTGAATTTACCGAATACTTTATTGGCCCAAAGCGTTCGTTTTGTGATGGCAGCTGCTATATTTTTGTCTTACGGCCTTCAATTTTACGTGCCAATGAACATTGTTTGGCCCTACGTGAAATCTAAGCTTTCATCGGAAAATGCTTTAAAGCACGGCGAGGCGATGACCCGTATCGCCCTAATTTCAATAACGTTTTTGGCTGCAGCGATGATTCCGAACTTGAGTGGAATAATTTCATTGGTTGGCGCGTTCAGTAGCTCTGCGCTCGCACTCATATTTCCACCACTGATCGAGATCATGACGTTTTGGCCGGATCAGCTCGGGAAAAACGATTGGAAATTTTGGAAAGATATCGTCATTATTGTTTTCGGTGTAACAGGTTTCCTGTTCGGTACTTATACTAGTATAGAGAATATATTAATAAAATCTTAA
Protein
MSDEKQPLLTGPNSPENVIEIQSTEPVINDDGPSTEKTKGDYCPASERCLEYPTSNFDTMIHLLKGNIGTGILAMPDAFKNAGLIFGVFCTLIMGAMCTHCMHILVQCSHELCIRSEKPALSFSEVIEDSFLSGPIVFRPYAKKMRALVNIFLVITQLGFCCVYFLFVATNLQDTMHLFRINLSVHLYLTLLFPLIVALAMVKNLKYLTPVSLVASIMTAWGLAITFYYILQDLPHSKEVNPIATWHQLPLYFGTAIYAFEGIGVVLPLENNMKTPEDFGGWNGVLNTGMVIVAALYTAVGFFGYLKYGDSVQGSITLNLPNTLLAQSVRFVMAAAIFLSYGLQFYVPMNIVWPYVKSKLSSENALKHGEAMTRIALISITFLAAAMIPNLSGIISLVGAFSSSALALIFPPLIEIMTFWPDQLGKNDWKFWKDIVIIVFGVTGFLFGTYTSIENILIKS
Summary
Uniprot
A0A212FG41
A0A1E1WB53
A0A2H1V8R1
A0A194PHM5
A0A1L8DEK2
A0A1L8DDY5
+ More
A0A1L8DEE0 Q17IV0 Q17IU9 A0A2M4CM91 Q7Q4M8 W5JUL6 B0W8T6 A0A182IZE0 A0A1S4GYE9 A0A182UEC3 A0A182IBW8 A0A182Y5J6 A0A182L059 A0A2M4AR61 A0A2M4BLC6 A0A182X157 A0A182P3B0 A0A2M3Z2T8 A0A182F5N3 A0A2M4ARN7 A0A084VL57 A0A182QYJ3 A0A1Q3F748 A0A182K3F4 A0A023EUN0 A0A182UUY5 A0A1Q3F6V9 A0A182GMW5 A0A1Q3F711 A0A1Q3F7F5 A0A182GU67 A0A182NUV6 A0A1B0CRP7 A0A336K4F4 A0A1Y0AWM8 A0A336K341 A0A182RRJ9 A0A139WJU4 A0A067R5W4 A0A182MQR8 A0A2J7RC29 A0A1J1IQZ7 A0A1Y1KGU6 A0A1Y1KLT8 A0A0T6AX94 A0A182WHH5 A0A1I8NFE8 A0A1B6C7H6 A0A0A1X319 A0A0A1WV48 W8BQQ7 A0A1B6CS67 W8ASK8 A0A2P8XI06 W8B467 A0A1I8QAY1 A0A1W4X1T4 A0A1W4WS54 A0A034V7C8 A0A1W4W7Y5 A0A1W4VWZ2 A0A0K8UAA4 A0A1B6GKB0 A0A1B6GMS5 A0A1B6EQ50 A0A1W4W990 A0A1W4W8T2 A0A1A9WM38 A0A0Q5WN47 A0A0Q5WBI4 A0A0Q5WC18 B3N760 Q7KTI1 E8NH21 A0A0R1DKS4 A0A0J9QZG7 B5DHP7 B4GJT5 A0A0R1DLQ3 A0A0R1DQ70 Q8MSR2 A0A0J9QYN8 A0A0J9QY85 Q9VLM3 A0A0R1DS10 A0A0J9TIW2 A0A0P9BN55 A0A0R3NRG6 A0A0R3NRF3 Q9VLM4 B4NWY1 A0A0R1DS44 A0A0K8W200 A0A3B0KDA6
A0A1L8DEE0 Q17IV0 Q17IU9 A0A2M4CM91 Q7Q4M8 W5JUL6 B0W8T6 A0A182IZE0 A0A1S4GYE9 A0A182UEC3 A0A182IBW8 A0A182Y5J6 A0A182L059 A0A2M4AR61 A0A2M4BLC6 A0A182X157 A0A182P3B0 A0A2M3Z2T8 A0A182F5N3 A0A2M4ARN7 A0A084VL57 A0A182QYJ3 A0A1Q3F748 A0A182K3F4 A0A023EUN0 A0A182UUY5 A0A1Q3F6V9 A0A182GMW5 A0A1Q3F711 A0A1Q3F7F5 A0A182GU67 A0A182NUV6 A0A1B0CRP7 A0A336K4F4 A0A1Y0AWM8 A0A336K341 A0A182RRJ9 A0A139WJU4 A0A067R5W4 A0A182MQR8 A0A2J7RC29 A0A1J1IQZ7 A0A1Y1KGU6 A0A1Y1KLT8 A0A0T6AX94 A0A182WHH5 A0A1I8NFE8 A0A1B6C7H6 A0A0A1X319 A0A0A1WV48 W8BQQ7 A0A1B6CS67 W8ASK8 A0A2P8XI06 W8B467 A0A1I8QAY1 A0A1W4X1T4 A0A1W4WS54 A0A034V7C8 A0A1W4W7Y5 A0A1W4VWZ2 A0A0K8UAA4 A0A1B6GKB0 A0A1B6GMS5 A0A1B6EQ50 A0A1W4W990 A0A1W4W8T2 A0A1A9WM38 A0A0Q5WN47 A0A0Q5WBI4 A0A0Q5WC18 B3N760 Q7KTI1 E8NH21 A0A0R1DKS4 A0A0J9QZG7 B5DHP7 B4GJT5 A0A0R1DLQ3 A0A0R1DQ70 Q8MSR2 A0A0J9QYN8 A0A0J9QY85 Q9VLM3 A0A0R1DS10 A0A0J9TIW2 A0A0P9BN55 A0A0R3NRG6 A0A0R3NRF3 Q9VLM4 B4NWY1 A0A0R1DS44 A0A0K8W200 A0A3B0KDA6
Pubmed
22118469
26354079
17510324
12364791
20920257
23761445
+ More
25244985 20966253 24438588 24945155 26483478 28341416 18362917 19820115 24845553 28004739 25315136 25830018 24495485 29403074 25348373 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085
25244985 20966253 24438588 24945155 26483478 28341416 18362917 19820115 24845553 28004739 25315136 25830018 24495485 29403074 25348373 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085
EMBL
AGBW02008720
OWR52705.1
GDQN01006832
JAT84222.1
ODYU01001259
SOQ37218.1
+ More
KQ459603 KPI92817.1 GFDF01009195 JAV04889.1 GFDF01009550 JAV04534.1 GFDF01009329 JAV04755.1 CH477236 EAT46644.1 EAT46643.1 GGFL01002225 MBW66403.1 AAAB01008964 EAA12852.4 ADMH02000447 ETN66434.1 DS231860 EDS39315.1 APCN01000752 GGFK01009939 MBW43260.1 GGFJ01004725 MBW53866.1 GGFM01002052 MBW22803.1 GGFK01010145 MBW43466.1 ATLV01014417 KE524970 KFB38701.1 AXCN02000747 GFDL01011696 JAV23349.1 GAPW01000931 JAC12667.1 GFDL01011749 JAV23296.1 JXUM01075159 KQ562872 KXJ74957.1 GFDL01011695 JAV23350.1 GFDL01011549 JAV23496.1 JXUM01088613 KQ563713 KXJ73450.1 AJWK01025074 AJWK01025075 UFQS01000097 UFQT01000097 SSW99387.1 SSX19767.1 KY921806 ART29395.1 SSW99386.1 SSX19766.1 KQ971338 KYB28192.1 KK852674 KDR18729.1 AXCM01001953 NEVH01005887 PNF38368.1 CVRI01000058 CRL02568.1 GEZM01086058 JAV59808.1 GEZM01086059 JAV59807.1 LJIG01022599 KRT79718.1 GEDC01027855 JAS09443.1 GBXI01008578 JAD05714.1 GBXI01017532 GBXI01011781 JAC96759.1 JAD02511.1 GAMC01014701 JAB91854.1 GEDC01020919 JAS16379.1 GAMC01014700 JAB91855.1 PYGN01002063 PSN31633.1 GAMC01014702 JAB91853.1 GAKP01021297 JAC37655.1 GDHF01032132 GDHF01028702 GDHF01003172 JAI20182.1 JAI23612.1 JAI49142.1 GECZ01029278 GECZ01027150 GECZ01006892 JAS40491.1 JAS42619.1 JAS62877.1 GECZ01029306 GECZ01024403 GECZ01023598 GECZ01022704 GECZ01015813 GECZ01006021 JAS40463.1 JAS45366.1 JAS46171.1 JAS47065.1 JAS53956.1 JAS63748.1 GECZ01029704 GECZ01028042 GECZ01016209 GECZ01002500 JAS40065.1 JAS41727.1 JAS53560.1 JAS67269.1 CH954177 KQS70770.1 KQS70771.1 KQS70772.1 EDV59356.1 AE014134 AAF52661.1 AAN10663.1 AAN10664.1 ADV36973.1 ADV36974.1 BT125875 ADV15454.1 CM000157 KRJ97871.1 CM002910 KMY89069.1 CH379058 EDY69824.1 CH479184 EDW36901.1 KRJ97870.1 KRJ97874.1 AY118648 AAM50017.1 AAS64738.1 KMY89073.1 KMY89072.1 AAF52662.2 KRJ97872.1 KMY89070.1 CH902620 KPU73250.1 KRT03594.1 KRT03593.1 BT125876 AAF52663.1 ADV15455.1 ADV36975.1 EDW88511.1 KRJ97873.1 GDHF01032920 GDHF01007449 JAI19394.1 JAI44865.1 OUUW01000010 SPP86230.1
KQ459603 KPI92817.1 GFDF01009195 JAV04889.1 GFDF01009550 JAV04534.1 GFDF01009329 JAV04755.1 CH477236 EAT46644.1 EAT46643.1 GGFL01002225 MBW66403.1 AAAB01008964 EAA12852.4 ADMH02000447 ETN66434.1 DS231860 EDS39315.1 APCN01000752 GGFK01009939 MBW43260.1 GGFJ01004725 MBW53866.1 GGFM01002052 MBW22803.1 GGFK01010145 MBW43466.1 ATLV01014417 KE524970 KFB38701.1 AXCN02000747 GFDL01011696 JAV23349.1 GAPW01000931 JAC12667.1 GFDL01011749 JAV23296.1 JXUM01075159 KQ562872 KXJ74957.1 GFDL01011695 JAV23350.1 GFDL01011549 JAV23496.1 JXUM01088613 KQ563713 KXJ73450.1 AJWK01025074 AJWK01025075 UFQS01000097 UFQT01000097 SSW99387.1 SSX19767.1 KY921806 ART29395.1 SSW99386.1 SSX19766.1 KQ971338 KYB28192.1 KK852674 KDR18729.1 AXCM01001953 NEVH01005887 PNF38368.1 CVRI01000058 CRL02568.1 GEZM01086058 JAV59808.1 GEZM01086059 JAV59807.1 LJIG01022599 KRT79718.1 GEDC01027855 JAS09443.1 GBXI01008578 JAD05714.1 GBXI01017532 GBXI01011781 JAC96759.1 JAD02511.1 GAMC01014701 JAB91854.1 GEDC01020919 JAS16379.1 GAMC01014700 JAB91855.1 PYGN01002063 PSN31633.1 GAMC01014702 JAB91853.1 GAKP01021297 JAC37655.1 GDHF01032132 GDHF01028702 GDHF01003172 JAI20182.1 JAI23612.1 JAI49142.1 GECZ01029278 GECZ01027150 GECZ01006892 JAS40491.1 JAS42619.1 JAS62877.1 GECZ01029306 GECZ01024403 GECZ01023598 GECZ01022704 GECZ01015813 GECZ01006021 JAS40463.1 JAS45366.1 JAS46171.1 JAS47065.1 JAS53956.1 JAS63748.1 GECZ01029704 GECZ01028042 GECZ01016209 GECZ01002500 JAS40065.1 JAS41727.1 JAS53560.1 JAS67269.1 CH954177 KQS70770.1 KQS70771.1 KQS70772.1 EDV59356.1 AE014134 AAF52661.1 AAN10663.1 AAN10664.1 ADV36973.1 ADV36974.1 BT125875 ADV15454.1 CM000157 KRJ97871.1 CM002910 KMY89069.1 CH379058 EDY69824.1 CH479184 EDW36901.1 KRJ97870.1 KRJ97874.1 AY118648 AAM50017.1 AAS64738.1 KMY89073.1 KMY89072.1 AAF52662.2 KRJ97872.1 KMY89070.1 CH902620 KPU73250.1 KRT03594.1 KRT03593.1 BT125876 AAF52663.1 ADV15455.1 ADV36975.1 EDW88511.1 KRJ97873.1 GDHF01032920 GDHF01007449 JAI19394.1 JAI44865.1 OUUW01000010 SPP86230.1
Proteomes
UP000007151
UP000053268
UP000008820
UP000007062
UP000000673
UP000002320
+ More
UP000075880 UP000075902 UP000075840 UP000076408 UP000075882 UP000076407 UP000075885 UP000069272 UP000030765 UP000075886 UP000075881 UP000075903 UP000069940 UP000249989 UP000075884 UP000092461 UP000075900 UP000007266 UP000027135 UP000075883 UP000235965 UP000183832 UP000075920 UP000095301 UP000245037 UP000095300 UP000192223 UP000192221 UP000091820 UP000008711 UP000000803 UP000002282 UP000001819 UP000008744 UP000007801 UP000268350
UP000075880 UP000075902 UP000075840 UP000076408 UP000075882 UP000076407 UP000075885 UP000069272 UP000030765 UP000075886 UP000075881 UP000075903 UP000069940 UP000249989 UP000075884 UP000092461 UP000075900 UP000007266 UP000027135 UP000075883 UP000235965 UP000183832 UP000075920 UP000095301 UP000245037 UP000095300 UP000192223 UP000192221 UP000091820 UP000008711 UP000000803 UP000002282 UP000001819 UP000008744 UP000007801 UP000268350
Pfam
PF01490 Aa_trans
Interpro
IPR013057
AA_transpt_TM
ProteinModelPortal
A0A212FG41
A0A1E1WB53
A0A2H1V8R1
A0A194PHM5
A0A1L8DEK2
A0A1L8DDY5
+ More
A0A1L8DEE0 Q17IV0 Q17IU9 A0A2M4CM91 Q7Q4M8 W5JUL6 B0W8T6 A0A182IZE0 A0A1S4GYE9 A0A182UEC3 A0A182IBW8 A0A182Y5J6 A0A182L059 A0A2M4AR61 A0A2M4BLC6 A0A182X157 A0A182P3B0 A0A2M3Z2T8 A0A182F5N3 A0A2M4ARN7 A0A084VL57 A0A182QYJ3 A0A1Q3F748 A0A182K3F4 A0A023EUN0 A0A182UUY5 A0A1Q3F6V9 A0A182GMW5 A0A1Q3F711 A0A1Q3F7F5 A0A182GU67 A0A182NUV6 A0A1B0CRP7 A0A336K4F4 A0A1Y0AWM8 A0A336K341 A0A182RRJ9 A0A139WJU4 A0A067R5W4 A0A182MQR8 A0A2J7RC29 A0A1J1IQZ7 A0A1Y1KGU6 A0A1Y1KLT8 A0A0T6AX94 A0A182WHH5 A0A1I8NFE8 A0A1B6C7H6 A0A0A1X319 A0A0A1WV48 W8BQQ7 A0A1B6CS67 W8ASK8 A0A2P8XI06 W8B467 A0A1I8QAY1 A0A1W4X1T4 A0A1W4WS54 A0A034V7C8 A0A1W4W7Y5 A0A1W4VWZ2 A0A0K8UAA4 A0A1B6GKB0 A0A1B6GMS5 A0A1B6EQ50 A0A1W4W990 A0A1W4W8T2 A0A1A9WM38 A0A0Q5WN47 A0A0Q5WBI4 A0A0Q5WC18 B3N760 Q7KTI1 E8NH21 A0A0R1DKS4 A0A0J9QZG7 B5DHP7 B4GJT5 A0A0R1DLQ3 A0A0R1DQ70 Q8MSR2 A0A0J9QYN8 A0A0J9QY85 Q9VLM3 A0A0R1DS10 A0A0J9TIW2 A0A0P9BN55 A0A0R3NRG6 A0A0R3NRF3 Q9VLM4 B4NWY1 A0A0R1DS44 A0A0K8W200 A0A3B0KDA6
A0A1L8DEE0 Q17IV0 Q17IU9 A0A2M4CM91 Q7Q4M8 W5JUL6 B0W8T6 A0A182IZE0 A0A1S4GYE9 A0A182UEC3 A0A182IBW8 A0A182Y5J6 A0A182L059 A0A2M4AR61 A0A2M4BLC6 A0A182X157 A0A182P3B0 A0A2M3Z2T8 A0A182F5N3 A0A2M4ARN7 A0A084VL57 A0A182QYJ3 A0A1Q3F748 A0A182K3F4 A0A023EUN0 A0A182UUY5 A0A1Q3F6V9 A0A182GMW5 A0A1Q3F711 A0A1Q3F7F5 A0A182GU67 A0A182NUV6 A0A1B0CRP7 A0A336K4F4 A0A1Y0AWM8 A0A336K341 A0A182RRJ9 A0A139WJU4 A0A067R5W4 A0A182MQR8 A0A2J7RC29 A0A1J1IQZ7 A0A1Y1KGU6 A0A1Y1KLT8 A0A0T6AX94 A0A182WHH5 A0A1I8NFE8 A0A1B6C7H6 A0A0A1X319 A0A0A1WV48 W8BQQ7 A0A1B6CS67 W8ASK8 A0A2P8XI06 W8B467 A0A1I8QAY1 A0A1W4X1T4 A0A1W4WS54 A0A034V7C8 A0A1W4W7Y5 A0A1W4VWZ2 A0A0K8UAA4 A0A1B6GKB0 A0A1B6GMS5 A0A1B6EQ50 A0A1W4W990 A0A1W4W8T2 A0A1A9WM38 A0A0Q5WN47 A0A0Q5WBI4 A0A0Q5WC18 B3N760 Q7KTI1 E8NH21 A0A0R1DKS4 A0A0J9QZG7 B5DHP7 B4GJT5 A0A0R1DLQ3 A0A0R1DQ70 Q8MSR2 A0A0J9QYN8 A0A0J9QY85 Q9VLM3 A0A0R1DS10 A0A0J9TIW2 A0A0P9BN55 A0A0R3NRG6 A0A0R3NRF3 Q9VLM4 B4NWY1 A0A0R1DS44 A0A0K8W200 A0A3B0KDA6
Ontologies
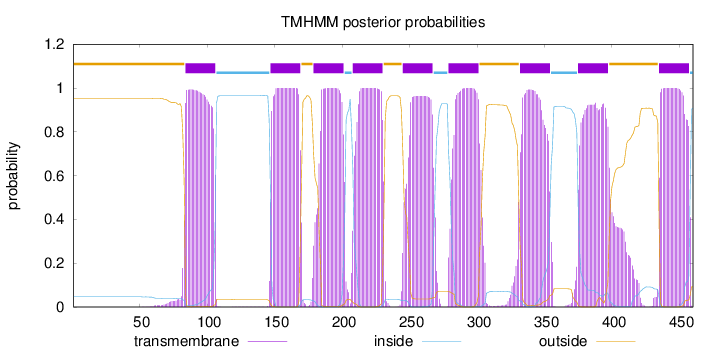
Topology
Length:
460
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
204.87802
Exp number, first 60 AAs:
0.01219
Total prob of N-in:
0.04677
outside
1 - 83
TMhelix
84 - 106
inside
107 - 146
TMhelix
147 - 169
outside
170 - 178
TMhelix
179 - 201
inside
202 - 207
TMhelix
208 - 230
outside
231 - 244
TMhelix
245 - 267
inside
268 - 278
TMhelix
279 - 301
outside
302 - 331
TMhelix
332 - 354
inside
355 - 374
TMhelix
375 - 397
outside
398 - 434
TMhelix
435 - 457
inside
458 - 460
Population Genetic Test Statistics
Pi
0.876469
Theta
5.67485
Tajima's D
-2.487546
CLR
596.992769
CSRT
0.000199990000499975
Interpretation
Possibly Positive selection
