Gene
KWMTBOMO07241
Annotation
PREDICTED:_TBC1_domain_family_member_23_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.102
Sequence
CDS
ATGGCAGGAGACGACGATGATACTTGGTTGATAGAACTAGAGTCTGCTCTTCTCGATGGATGTACCGCACAAGAAATTAATGCATTAACGAAAGGAAAAAAAATACCTGTTAGCTTACGTCCTGATGTGTGGTTGTTATGTTTGAATTGTCAAGATGCTGGAAACCAATTGCTTTTGTTTGATGAGATATTTGACTTACCTAATCAGAATGAATTAAGAGATGATGTAAAAAAGTTTGTTGAAAAACTTGGCAATGAAGAAGATGATAAATTAGCAGTTATATCTGATGTTGAGTCAATAATCACATTTTATTGCAAATCGAAGAACACAACTTACACTTCAAACAATGGTTGGATTGATATTTTATTACCTTTGTTAAGTTTGAAGCTACCACGGTCAGATACTTATAATTTGTTTGAAAGGATTTTAAAATTGTATACACCAAAGGGCTGTTCAAAAAATGGAGTACCTTGCCATATATTAAGGTTGATACTTCAATACCATGATCCAGAACTTTGTTCATTTTTGGATACAAAAAGAATCACTCCAGAACAGTATTGTTCACCATGGTTGAAGTCATTATTTGCTGGTACATGTAGTTTGGATGTAGCTTTATACATGTGGGATTTGTATTTTCAAAGGTCAGATCCGTTCTTCATATTTTTTCTATGCTTAATTATGATCATTAATGCCAGAGAACAATTACTACAGATGAAAAATGATGATAAAACAGCCATTATACAAATTCTTACCAATATGCCTGCAGATTTGGAAGCAAATGATGTTTCGGATTTCTGTTCATTGGCTCATTACTACTCATTAAAAACACCTCAGTCTTTCAGAGATGATGTCTTAGAGGTCTTATTTTCAGAAAGTGAATTGGAAATAAATTCAAGACTGTACTCACAAGCTTTATGTCTGCCTGTAGCAGTTCATGAATTAATTGAAAGTGCAACATTAGAATTGGCAAATTCAGATAGTGTGAAATTCTTTTTAGTTGATTGTCGACCAACAGATCAATATAATGCAGGTCATTTATCCACCGCATTTCATCTAGATTGTAATTTGATGTTGCAAGAACCTGCAGCTTTTAATACAGCCGTGCAAGGACTATTAAATGCTCAAAGACAAGCCTTAGCTGCTGGATCACTTGCTGCTGGAGAACATCTATGTTTTGTCGGATCAGGTAGAACTGAAGAAGATAGTTATGCTCATATGGTGGTTGCATCATTTCTGAAGAGAAATACTAAACATGTTTCTATGCTTGATGGTGGCTTTGTGGCAATTCATGATTATTTTGGGCCTCATATGACAGATTGCCTAGAGGAACACAATGCATCAGCATGTCTTGTTTGTGTTCCAAATAATACCTCTGTTGGAATGAGTAAAAACGTAGCCAGTGCAACCAAAACTAGTGAGAATATGATTACTAAACAATTGTTTAGCAAATTGTCATCTGCCATGAAAGCTAAAAGTCAAGAAGTCAAAGGGAAATTGATAGAGTACATAGTTAACCCAAATTCCACATACAATAATAGTGAATGGCATGTGTCTGCAAATGATAGAAAAGAGCAAAGGTATAGAAATGTACCACCTGTATTTAGCATAAATGATGAAGATGAAGACTCACGGTCTGATAGTTTACGTCATGATCTTACACTTTCTGATTCTATCGAAGAAATAGTTAATGTGCAGTCGTTTCTTAAGGAACCAAATACTATTGATAATTTTCAATGCCAGGAAGTATTACTTAATGGATATATGCATGATTCTTATATAGTTGTAACAGAAACACACTTAATTGTTTTACGTGACATTCCAAATAAACGAGGCCTAGCAAAGATTATGTCTAGGAGACCTCTCTCTACTATAGTTAAAATAACAGCTAAGAAGAGGCATCCAGAATTAATTACATTTAAGTATGGTATTCCAGATGGTGATAATTTACTAATTAAAGATATGGATAGATTTCTTATTCCAAATGCCTCTGTAGCTACTAAAGTTGTATCAAATCAAATTGTACAACAGTTGGATAACAAATTACAGTGA
Protein
MAGDDDDTWLIELESALLDGCTAQEINALTKGKKIPVSLRPDVWLLCLNCQDAGNQLLLFDEIFDLPNQNELRDDVKKFVEKLGNEEDDKLAVISDVESIITFYCKSKNTTYTSNNGWIDILLPLLSLKLPRSDTYNLFERILKLYTPKGCSKNGVPCHILRLILQYHDPELCSFLDTKRITPEQYCSPWLKSLFAGTCSLDVALYMWDLYFQRSDPFFIFFLCLIMIINAREQLLQMKNDDKTAIIQILTNMPADLEANDVSDFCSLAHYYSLKTPQSFRDDVLEVLFSESELEINSRLYSQALCLPVAVHELIESATLELANSDSVKFFLVDCRPTDQYNAGHLSTAFHLDCNLMLQEPAAFNTAVQGLLNAQRQALAAGSLAAGEHLCFVGSGRTEEDSYAHMVVASFLKRNTKHVSMLDGGFVAIHDYFGPHMTDCLEEHNASACLVCVPNNTSVGMSKNVASATKTSENMITKQLFSKLSSAMKAKSQEVKGKLIEYIVNPNSTYNNSEWHVSANDRKEQRYRNVPPVFSINDEDEDSRSDSLRHDLTLSDSIEEIVNVQSFLKEPNTIDNFQCQEVLLNGYMHDSYIVVTETHLIVLRDIPNKRGLAKIMSRRPLSTIVKITAKKRHPELITFKYGIPDGDNLLIKDMDRFLIPNASVATKVVSNQIVQQLDNKLQ
Summary
Uniprot
A0A1E1WI32
A0A2H1VAU5
A0A2A4IX48
A0A2W1BUR8
A0A194R0C2
A0A212FG48
+ More
A0A0L7KT23 V5I9S9 A0A1Y1N1Q6 A0A182GSQ8 U4UNE2 A0A2J7PK67 Q16FF8 A0A1L8DAR7 A0A0K8TQN7 N6TED7 A0A0A1WNE6 D2A1Y0 A0A1B0DEN2 U5EYH8 A0A0K8UD91 A0A034WH11 W8C0R0 A0A0T6AYD6 A0A1A9UTW2 A0A067RCP6 A0A1W4XAY4 A0A1Q3F5I5 A0A1A9YR49 A0A1Q3F557 A0A1B0BDR8 A0A1B0CW71 B4KEU1 B0W0C9 A0A1A9Z5X4 A0A1I8MF47 T1PFM3 A0A0L0CIF8 B4JDE8 A0A1Q3F4Y6 B4LU39 A0A1B0FMP7 A0A1Q3F5B4 A0A0M4EPP2 B3MN91 A0A1A9WN26 A0A0Q9XFT4 A0A3B0JC25 A0A2J7PK73 Q29LH8 Q9VPW9 Q8MRS6 A0A1Q3F536 A0A1Q3F544 A0A1I8Q808 B4Q6M9 A0A1W4VY50 B3N8B9 A0A1W4WAF7 A0A1S4GWS4 A0A182L2P9 B4P308 A0A182IEC4 A0A182X1E4 A0A182TGQ3 Q7Q378 A0A182PCW8 A0A182JR98 B4MZI8 A0A182W7L2 A0A182UYE2 A0A182XZR8 A0A182MX44 A0A1W4XC32 A0A182RJD8 A0A2M4ADZ8 A0A2M4BF45 A0A2M4ADY7 A0A2M4AE59 A0A2M4ADX9 A0A2M4BF19 A0A182SPB8 A0A2M4BFB8 A0A182QBN3 A0A182NTG6 K7IZS9 A0A182JGG3 A0A084WHF3 A0A232FEZ0 A0A0C9QYJ4 A0A195B9P5 A0A158NBI5 F4X8P0 A0A195DRA8 A0A195FCX6 E9IQE6 A0A0Q9WBJ9 A0A151IGY6 A0A026W4N5 E2B021
A0A0L7KT23 V5I9S9 A0A1Y1N1Q6 A0A182GSQ8 U4UNE2 A0A2J7PK67 Q16FF8 A0A1L8DAR7 A0A0K8TQN7 N6TED7 A0A0A1WNE6 D2A1Y0 A0A1B0DEN2 U5EYH8 A0A0K8UD91 A0A034WH11 W8C0R0 A0A0T6AYD6 A0A1A9UTW2 A0A067RCP6 A0A1W4XAY4 A0A1Q3F5I5 A0A1A9YR49 A0A1Q3F557 A0A1B0BDR8 A0A1B0CW71 B4KEU1 B0W0C9 A0A1A9Z5X4 A0A1I8MF47 T1PFM3 A0A0L0CIF8 B4JDE8 A0A1Q3F4Y6 B4LU39 A0A1B0FMP7 A0A1Q3F5B4 A0A0M4EPP2 B3MN91 A0A1A9WN26 A0A0Q9XFT4 A0A3B0JC25 A0A2J7PK73 Q29LH8 Q9VPW9 Q8MRS6 A0A1Q3F536 A0A1Q3F544 A0A1I8Q808 B4Q6M9 A0A1W4VY50 B3N8B9 A0A1W4WAF7 A0A1S4GWS4 A0A182L2P9 B4P308 A0A182IEC4 A0A182X1E4 A0A182TGQ3 Q7Q378 A0A182PCW8 A0A182JR98 B4MZI8 A0A182W7L2 A0A182UYE2 A0A182XZR8 A0A182MX44 A0A1W4XC32 A0A182RJD8 A0A2M4ADZ8 A0A2M4BF45 A0A2M4ADY7 A0A2M4AE59 A0A2M4ADX9 A0A2M4BF19 A0A182SPB8 A0A2M4BFB8 A0A182QBN3 A0A182NTG6 K7IZS9 A0A182JGG3 A0A084WHF3 A0A232FEZ0 A0A0C9QYJ4 A0A195B9P5 A0A158NBI5 F4X8P0 A0A195DRA8 A0A195FCX6 E9IQE6 A0A0Q9WBJ9 A0A151IGY6 A0A026W4N5 E2B021
Pubmed
28756777
26354079
22118469
26227816
28004739
26483478
+ More
23537049 17510324 26369729 25830018 18362917 19820115 25348373 24495485 24845553 17994087 25315136 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 12364791 20966253 17550304 25244985 20075255 24438588 28648823 21347285 21719571 21282665 24508170 30249741 20798317
23537049 17510324 26369729 25830018 18362917 19820115 25348373 24495485 24845553 17994087 25315136 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 12364791 20966253 17550304 25244985 20075255 24438588 28648823 21347285 21719571 21282665 24508170 30249741 20798317
EMBL
GDQN01004412
JAT86642.1
ODYU01001570
SOQ37968.1
NWSH01005736
PCG63998.1
+ More
KZ149911 PZC78111.1 KQ460883 KPJ11147.1 AGBW02008720 OWR52708.1 JTDY01005957 KOB66433.1 GALX01002405 JAB66061.1 GEZM01015241 GEZM01015240 JAV91824.1 JXUM01017635 KQ560488 KXJ82158.1 KB632375 ERL93983.1 NEVH01024940 PNF16727.1 CH478448 EAT32974.1 GFDF01010523 JAV03561.1 GDAI01000939 JAI16664.1 APGK01032481 KB740842 ENN78734.1 GBXI01014091 JAD00201.1 KQ971338 EFA02068.1 AJVK01005691 GANO01000289 JAB59582.1 GDHF01027989 JAI24325.1 GAKP01004096 JAC54856.1 GAMC01001278 JAC05278.1 LJIG01022522 KRT80134.1 KK852723 KDR17691.1 GFDL01012223 JAV22822.1 GFDL01012348 JAV22697.1 JXJN01012652 AJWK01031863 CH933807 EDW12991.1 DS231817 EDS39668.1 KA646703 AFP61332.1 JRES01000438 KNC31274.1 CH916368 EDW03318.1 GFDL01012420 JAV22625.1 CH940649 EDW65092.1 CCAG010018220 GFDL01012299 JAV22746.1 CP012523 ALC38582.1 CH902620 EDV31048.1 KRG03534.1 OUUW01000004 SPP79914.1 PNF16728.1 CH379060 EAL34067.2 AE014134 AAF51416.2 AY119443 AAM50097.1 GFDL01012371 JAV22674.1 GFDL01012422 JAV22623.1 CM000361 CM002910 EDX03295.1 KMY87422.1 CH954177 EDV57306.1 AAAB01008964 CM000157 EDW87215.1 APCN01005200 EAA12378.4 CH963920 EDW77773.1 AXCM01001882 GGFK01005690 MBW39011.1 GGFJ01002511 MBW51652.1 GGFK01005639 MBW38960.1 GGFK01005752 MBW39073.1 GGFK01005670 MBW38991.1 GGFJ01002509 MBW51650.1 GGFJ01002510 MBW51651.1 AXCN02000703 ATLV01023809 KE525346 KFB49647.1 NNAY01000319 OXU29255.1 GBYB01008851 GBYB01008852 GBYB01008854 JAG78618.1 JAG78619.1 JAG78621.1 KQ976540 KYM81251.1 ADTU01011113 GL888932 EGI57309.1 KQ980581 KYN15403.1 KQ981673 KYN38241.1 GL764741 EFZ17207.1 KRF82009.1 KQ977661 KYN00699.1 KK107453 QOIP01000004 EZA50556.1 RLU23448.1 GL444349 EFN60966.1
KZ149911 PZC78111.1 KQ460883 KPJ11147.1 AGBW02008720 OWR52708.1 JTDY01005957 KOB66433.1 GALX01002405 JAB66061.1 GEZM01015241 GEZM01015240 JAV91824.1 JXUM01017635 KQ560488 KXJ82158.1 KB632375 ERL93983.1 NEVH01024940 PNF16727.1 CH478448 EAT32974.1 GFDF01010523 JAV03561.1 GDAI01000939 JAI16664.1 APGK01032481 KB740842 ENN78734.1 GBXI01014091 JAD00201.1 KQ971338 EFA02068.1 AJVK01005691 GANO01000289 JAB59582.1 GDHF01027989 JAI24325.1 GAKP01004096 JAC54856.1 GAMC01001278 JAC05278.1 LJIG01022522 KRT80134.1 KK852723 KDR17691.1 GFDL01012223 JAV22822.1 GFDL01012348 JAV22697.1 JXJN01012652 AJWK01031863 CH933807 EDW12991.1 DS231817 EDS39668.1 KA646703 AFP61332.1 JRES01000438 KNC31274.1 CH916368 EDW03318.1 GFDL01012420 JAV22625.1 CH940649 EDW65092.1 CCAG010018220 GFDL01012299 JAV22746.1 CP012523 ALC38582.1 CH902620 EDV31048.1 KRG03534.1 OUUW01000004 SPP79914.1 PNF16728.1 CH379060 EAL34067.2 AE014134 AAF51416.2 AY119443 AAM50097.1 GFDL01012371 JAV22674.1 GFDL01012422 JAV22623.1 CM000361 CM002910 EDX03295.1 KMY87422.1 CH954177 EDV57306.1 AAAB01008964 CM000157 EDW87215.1 APCN01005200 EAA12378.4 CH963920 EDW77773.1 AXCM01001882 GGFK01005690 MBW39011.1 GGFJ01002511 MBW51652.1 GGFK01005639 MBW38960.1 GGFK01005752 MBW39073.1 GGFK01005670 MBW38991.1 GGFJ01002509 MBW51650.1 GGFJ01002510 MBW51651.1 AXCN02000703 ATLV01023809 KE525346 KFB49647.1 NNAY01000319 OXU29255.1 GBYB01008851 GBYB01008852 GBYB01008854 JAG78618.1 JAG78619.1 JAG78621.1 KQ976540 KYM81251.1 ADTU01011113 GL888932 EGI57309.1 KQ980581 KYN15403.1 KQ981673 KYN38241.1 GL764741 EFZ17207.1 KRF82009.1 KQ977661 KYN00699.1 KK107453 QOIP01000004 EZA50556.1 RLU23448.1 GL444349 EFN60966.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000037510
UP000069940
UP000249989
+ More
UP000030742 UP000235965 UP000008820 UP000019118 UP000007266 UP000092462 UP000078200 UP000027135 UP000192223 UP000092443 UP000092460 UP000092461 UP000009192 UP000002320 UP000092445 UP000095301 UP000037069 UP000001070 UP000008792 UP000092444 UP000092553 UP000007801 UP000091820 UP000268350 UP000001819 UP000000803 UP000095300 UP000000304 UP000192221 UP000008711 UP000075882 UP000002282 UP000075840 UP000076407 UP000075902 UP000007062 UP000075885 UP000075881 UP000007798 UP000075920 UP000075903 UP000076408 UP000075883 UP000075900 UP000075901 UP000075886 UP000075884 UP000002358 UP000075880 UP000030765 UP000215335 UP000078540 UP000005205 UP000007755 UP000078492 UP000078541 UP000078542 UP000053097 UP000279307 UP000000311
UP000030742 UP000235965 UP000008820 UP000019118 UP000007266 UP000092462 UP000078200 UP000027135 UP000192223 UP000092443 UP000092460 UP000092461 UP000009192 UP000002320 UP000092445 UP000095301 UP000037069 UP000001070 UP000008792 UP000092444 UP000092553 UP000007801 UP000091820 UP000268350 UP000001819 UP000000803 UP000095300 UP000000304 UP000192221 UP000008711 UP000075882 UP000002282 UP000075840 UP000076407 UP000075902 UP000007062 UP000075885 UP000075881 UP000007798 UP000075920 UP000075903 UP000076408 UP000075883 UP000075900 UP000075901 UP000075886 UP000075884 UP000002358 UP000075880 UP000030765 UP000215335 UP000078540 UP000005205 UP000007755 UP000078492 UP000078541 UP000078542 UP000053097 UP000279307 UP000000311
Interpro
IPR001763
Rhodanese-like_dom
+ More
IPR035969 Rab-GTPase_TBC_sf
IPR039755 TBC1D23
IPR036873 Rhodanese-like_dom_sf
IPR000195 Rab-GTPase-TBC_dom
IPR013783 Ig-like_fold
IPR014756 Ig_E-set
IPR032640 AMPK1_CBM
IPR037256 ASC_dom_sf
IPR036869 J_dom_sf
IPR018253 DnaJ_domain_CS
IPR013766 Thioredoxin_domain
IPR036249 Thioredoxin-like_sf
IPR001623 DnaJ_domain
IPR035969 Rab-GTPase_TBC_sf
IPR039755 TBC1D23
IPR036873 Rhodanese-like_dom_sf
IPR000195 Rab-GTPase-TBC_dom
IPR013783 Ig-like_fold
IPR014756 Ig_E-set
IPR032640 AMPK1_CBM
IPR037256 ASC_dom_sf
IPR036869 J_dom_sf
IPR018253 DnaJ_domain_CS
IPR013766 Thioredoxin_domain
IPR036249 Thioredoxin-like_sf
IPR001623 DnaJ_domain
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A1E1WI32
A0A2H1VAU5
A0A2A4IX48
A0A2W1BUR8
A0A194R0C2
A0A212FG48
+ More
A0A0L7KT23 V5I9S9 A0A1Y1N1Q6 A0A182GSQ8 U4UNE2 A0A2J7PK67 Q16FF8 A0A1L8DAR7 A0A0K8TQN7 N6TED7 A0A0A1WNE6 D2A1Y0 A0A1B0DEN2 U5EYH8 A0A0K8UD91 A0A034WH11 W8C0R0 A0A0T6AYD6 A0A1A9UTW2 A0A067RCP6 A0A1W4XAY4 A0A1Q3F5I5 A0A1A9YR49 A0A1Q3F557 A0A1B0BDR8 A0A1B0CW71 B4KEU1 B0W0C9 A0A1A9Z5X4 A0A1I8MF47 T1PFM3 A0A0L0CIF8 B4JDE8 A0A1Q3F4Y6 B4LU39 A0A1B0FMP7 A0A1Q3F5B4 A0A0M4EPP2 B3MN91 A0A1A9WN26 A0A0Q9XFT4 A0A3B0JC25 A0A2J7PK73 Q29LH8 Q9VPW9 Q8MRS6 A0A1Q3F536 A0A1Q3F544 A0A1I8Q808 B4Q6M9 A0A1W4VY50 B3N8B9 A0A1W4WAF7 A0A1S4GWS4 A0A182L2P9 B4P308 A0A182IEC4 A0A182X1E4 A0A182TGQ3 Q7Q378 A0A182PCW8 A0A182JR98 B4MZI8 A0A182W7L2 A0A182UYE2 A0A182XZR8 A0A182MX44 A0A1W4XC32 A0A182RJD8 A0A2M4ADZ8 A0A2M4BF45 A0A2M4ADY7 A0A2M4AE59 A0A2M4ADX9 A0A2M4BF19 A0A182SPB8 A0A2M4BFB8 A0A182QBN3 A0A182NTG6 K7IZS9 A0A182JGG3 A0A084WHF3 A0A232FEZ0 A0A0C9QYJ4 A0A195B9P5 A0A158NBI5 F4X8P0 A0A195DRA8 A0A195FCX6 E9IQE6 A0A0Q9WBJ9 A0A151IGY6 A0A026W4N5 E2B021
A0A0L7KT23 V5I9S9 A0A1Y1N1Q6 A0A182GSQ8 U4UNE2 A0A2J7PK67 Q16FF8 A0A1L8DAR7 A0A0K8TQN7 N6TED7 A0A0A1WNE6 D2A1Y0 A0A1B0DEN2 U5EYH8 A0A0K8UD91 A0A034WH11 W8C0R0 A0A0T6AYD6 A0A1A9UTW2 A0A067RCP6 A0A1W4XAY4 A0A1Q3F5I5 A0A1A9YR49 A0A1Q3F557 A0A1B0BDR8 A0A1B0CW71 B4KEU1 B0W0C9 A0A1A9Z5X4 A0A1I8MF47 T1PFM3 A0A0L0CIF8 B4JDE8 A0A1Q3F4Y6 B4LU39 A0A1B0FMP7 A0A1Q3F5B4 A0A0M4EPP2 B3MN91 A0A1A9WN26 A0A0Q9XFT4 A0A3B0JC25 A0A2J7PK73 Q29LH8 Q9VPW9 Q8MRS6 A0A1Q3F536 A0A1Q3F544 A0A1I8Q808 B4Q6M9 A0A1W4VY50 B3N8B9 A0A1W4WAF7 A0A1S4GWS4 A0A182L2P9 B4P308 A0A182IEC4 A0A182X1E4 A0A182TGQ3 Q7Q378 A0A182PCW8 A0A182JR98 B4MZI8 A0A182W7L2 A0A182UYE2 A0A182XZR8 A0A182MX44 A0A1W4XC32 A0A182RJD8 A0A2M4ADZ8 A0A2M4BF45 A0A2M4ADY7 A0A2M4AE59 A0A2M4ADX9 A0A2M4BF19 A0A182SPB8 A0A2M4BFB8 A0A182QBN3 A0A182NTG6 K7IZS9 A0A182JGG3 A0A084WHF3 A0A232FEZ0 A0A0C9QYJ4 A0A195B9P5 A0A158NBI5 F4X8P0 A0A195DRA8 A0A195FCX6 E9IQE6 A0A0Q9WBJ9 A0A151IGY6 A0A026W4N5 E2B021
Ontologies
PANTHER
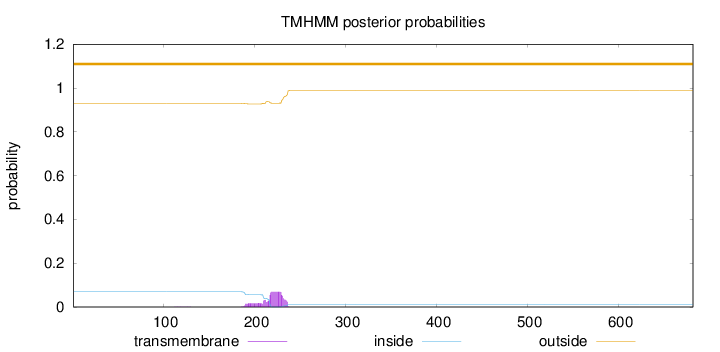
Topology
Length:
682
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.69829
Exp number, first 60 AAs:
0.00039
Total prob of N-in:
0.07074
outside
1 - 682
Population Genetic Test Statistics
Pi
0.622958
Theta
4.20324
Tajima's D
-2.339852
CLR
954.244596
CSRT
0.00124993750312484
Interpretation
Possibly Positive selection
