Gene
KWMTBOMO07236 Validated by peptides from experiments
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.205
Sequence
CDS
ATGGGAACAATGAAATTTTCGGTTTTGTGGATATTCATAGCATACCAACTGATTTATGTGAATAACGCATACAAAATAGATTTACTTGATGACTTAACGTCCGTTACTGACGATACAGAGAACGTTTTGGAGGAAATCGAATCGCCCTCCCAAAATTATAGAAACGGCAAAATCTTGTGGACTTTTCCGGGAAACCTTACCGAGCCGTTTAAAAAAAATGGCGTGAACCCTACTAGTGAAGGTTCAAAACTAACTGAACGACAAAAACGTTTCCTATTGTGGTCGTATCCTCAGAGCCCTTTAGTTGACATGATAATGCAAACGATGGCAAACAACAACTATTCACCCAAGAATTCACACGATCCTTTCGACTTTTTACGAGATTCGTATCCTCTGCCTAAAGGACATACAACACCACTTGAAGAGTATGATTATATCATAGTCGGAGGAGGAACGTCGGGGTCTGTTTTAGCTTCCCGCCTTACAGAGGACAAACCTAAGGCGACTGTATTAGTGATTGAAGCTGGTAAACCAGAGATGCTTATATCTGACATACCTGCATTAGCGCGGTATCTTCAGCTCACCGATTACGCGTGGCCGTACACTATGGAACACCAATCCGGTGTTTGCTTAGGTAGCGATGAACAGCGCTGCTACTGGCCGAGAGGAAAAGCTTTAGGTGGAAGTAGTGTCACTGATTTCATGTTATACACTCGAGGGCGACCACAAGACTGGGATAGAATCGCAGCCGATGGCAATTACGGATGGACCCATGACGAAGTTCTAAAATATTTCATGAAATCTGAAAGGTCAGAGTTAAGAAAATACAAGAACGCTTCTTATAGAGGACGAGATGGCGAACTTACAGTCGAAAATGTTCCGTTTAAAACCGGTTTAGTTGAAGCATTTCTGCAAGCCGGAAGAAACTTTGGTCATCCCACGGTTGATTACAATAACAACTCCCCAGATGAATTAGGTTTTGGCTATGTACAAACAACTACTACCAAGGGCCACCGATTGAGTACCGCCAAAGCATTTCTACATCCCCATAAGAGGAGAAAGAATTTACACATCTTAACTGAAGCTAGAGCTGGCAAAGTGATTATCGAACCACTTACTAAACGGGCCTATGCCGTTGAATACTTCAAAAATGGCGCTAAGTACACTGTTCGTTGCCGCAGAGAAGTAATTTTGGCTGCTGGCCCCATAGCTTCTCCGCAGTTACTAATGCTCTCAGGTATTGGGCCTCAAGAACATTTACAAACTCTGGGGATACCTGTATTGGCAAATTTACAAGTTGGAAGATCACTTTACGACCATATAGCTTTCCCTGGCGTTATTTTCAAATTGAATACCAGCAATGCTAGTCTTTTAGAACCTAAGGTAGCGACATTGCCAAATTTAATGCAATGGCTTCAATTCGGGGATGGACTATTAACTTCTCCTGGAATGATCGAAGCCTTAGGTTATATTAAAACTTCCGTTTCAAAGGATCCAGAACAAGTTCCGGATGTCGAGTTAATAAGCATGGGAACATCGATCAATGTAGATGGAGGTGGAGCTTTCAGAAAAAGCTGGCGAATCTCCGATAAAACATACGTAAAATCTTTCAGTTCTTTGAACGGTATGGACACGTGGTCAGCAATTCCAATGCTTTTACAACCAAAATCTACAGGTTATCTGGAATTAAGAGACACAAGTCCTTATTCTTTTCCAAAATTATACGGAAATTATTTAAAAGACCGTCAAGACATGGATACTTTATTGGAAGCTATAAAATACGTAATAAAATTGGGAGAATCGGAGCCTTTTAAAAAGTATGGAACAAGTCTGTTTCTGGCGGATTATCCATCATGCTCTGAACATACGCCTGGCTCAGATCCGTATTGGGAATGTGCTATCAGAACCATGGTTATATCTTTACATCACCAAGTAGGAACATGCAAGATGGGTCCACCTAGCGATTCTTATGCGGTTGTGGATCCAGAGCTAAGAGTGTACGGCATTGAAGGTCTTAGAGTGGTCGACGCAAGTATAATTCCGAAACCAATTTGTGCACACAGTACTGTACCTACAATAATGATTGCTGAAAAAGCCGCTGATATGATAAAACAAACATGGTCAAATGCTTCCGTTTAA
Protein
MGTMKFSVLWIFIAYQLIYVNNAYKIDLLDDLTSVTDDTENVLEEIESPSQNYRNGKILWTFPGNLTEPFKKNGVNPTSEGSKLTERQKRFLLWSYPQSPLVDMIMQTMANNNYSPKNSHDPFDFLRDSYPLPKGHTTPLEEYDYIIVGGGTSGSVLASRLTEDKPKATVLVIEAGKPEMLISDIPALARYLQLTDYAWPYTMEHQSGVCLGSDEQRCYWPRGKALGGSSVTDFMLYTRGRPQDWDRIAADGNYGWTHDEVLKYFMKSERSELRKYKNASYRGRDGELTVENVPFKTGLVEAFLQAGRNFGHPTVDYNNNSPDELGFGYVQTTTTKGHRLSTAKAFLHPHKRRKNLHILTEARAGKVIIEPLTKRAYAVEYFKNGAKYTVRCRREVILAAGPIASPQLLMLSGIGPQEHLQTLGIPVLANLQVGRSLYDHIAFPGVIFKLNTSNASLLEPKVATLPNLMQWLQFGDGLLTSPGMIEALGYIKTSVSKDPEQVPDVELISMGTSINVDGGGAFRKSWRISDKTYVKSFSSLNGMDTWSAIPMLLQPKSTGYLELRDTSPYSFPKLYGNYLKDRQDMDTLLEAIKYVIKLGESEPFKKYGTSLFLADYPSCSEHTPGSDPYWECAIRTMVISLHHQVGTCKMGPPSDSYAVVDPELRVYGIEGLRVVDASIIPKPICAHSTVPTIMIAEKAADMIKQTWSNASV
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
A0A212EHV5
A0A194R0C6
A0A194PID4
A0A2W1BSS3
A0A0L7KWS0
I4DN78
+ More
A0A2H1V5E5 A0A2H1W5U4 H9JTZ4 H9JTZ5 A0A2A4JIB2 A0A194RF91 A0A194QK24 H9JTZ7 A0A212FMN5 A0A194QIN4 A0A194RJ52 A0A2H1VEA7 E0VUA4 F4ZNI1 F4ZNI3 A0A2S2PXF2 A0A1S4F6C0 W4VSN1 Q17DV5 A0A2J7RBG4 J9JXJ8 Q16P03 A0A1S4FU90 A0A2H8TP19 A0A1B6DYE3 A0A1J1HR16 K7J274 A0A2S2PIK7 A0A1B0D7W4 A0A088AJU4 A0A182GDJ6 F4WR74 E9IKG7 A0A158NUR3 A0A182H0W9 A0A151J323 D2A3N4 W4VS94 A0A151K0E9 E1ZVM3 A0A0C9RGK1 A0A1B6JGN2 A0A3L8D5U9 A0A0L7LG80 A0A067RC53 E9IKG5 A0A194PXZ1 A0A194RJ41 E2A5J5 E9G8N9 A0A195FTT2 A0A158NUU8 A0A0U9HSH0 A0A310SX81 A0A195BAK2 A0A195E3M0 A0A151WMQ2 J9JWV7 A0A154PDJ9 A0A182F7P4 A0A154PD41 A0A1B6DKE4 A0A2K8FTL4 A0A182H3F8 A0A1W4XEG1 A0A0J7KKC5 A0A067QS81 F4WAD9 J9JM93 T1HCA7 A0A1Y1KXV7 A0A182LAT9 A0A2A4JHW7 A0A0J7KKD8 E2BJJ7 A0A084VGK6 A0A182H5D1 A0A182H3F7 A0A2J7PRD5 Q7PFS6 W5JAQ1 A0A0C9PS40 A0A2H8TLM1 A0A182WV76 B0WYR8 A0A182I8V1 K7J283 A0A0P5W9T8 A0A0P5PUH8 A0A182JLA4 A0A182TTM6 N6UKW6 A0A2J7QH94 E9IKF8
A0A2H1V5E5 A0A2H1W5U4 H9JTZ4 H9JTZ5 A0A2A4JIB2 A0A194RF91 A0A194QK24 H9JTZ7 A0A212FMN5 A0A194QIN4 A0A194RJ52 A0A2H1VEA7 E0VUA4 F4ZNI1 F4ZNI3 A0A2S2PXF2 A0A1S4F6C0 W4VSN1 Q17DV5 A0A2J7RBG4 J9JXJ8 Q16P03 A0A1S4FU90 A0A2H8TP19 A0A1B6DYE3 A0A1J1HR16 K7J274 A0A2S2PIK7 A0A1B0D7W4 A0A088AJU4 A0A182GDJ6 F4WR74 E9IKG7 A0A158NUR3 A0A182H0W9 A0A151J323 D2A3N4 W4VS94 A0A151K0E9 E1ZVM3 A0A0C9RGK1 A0A1B6JGN2 A0A3L8D5U9 A0A0L7LG80 A0A067RC53 E9IKG5 A0A194PXZ1 A0A194RJ41 E2A5J5 E9G8N9 A0A195FTT2 A0A158NUU8 A0A0U9HSH0 A0A310SX81 A0A195BAK2 A0A195E3M0 A0A151WMQ2 J9JWV7 A0A154PDJ9 A0A182F7P4 A0A154PD41 A0A1B6DKE4 A0A2K8FTL4 A0A182H3F8 A0A1W4XEG1 A0A0J7KKC5 A0A067QS81 F4WAD9 J9JM93 T1HCA7 A0A1Y1KXV7 A0A182LAT9 A0A2A4JHW7 A0A0J7KKD8 E2BJJ7 A0A084VGK6 A0A182H5D1 A0A182H3F7 A0A2J7PRD5 Q7PFS6 W5JAQ1 A0A0C9PS40 A0A2H8TLM1 A0A182WV76 B0WYR8 A0A182I8V1 K7J283 A0A0P5W9T8 A0A0P5PUH8 A0A182JLA4 A0A182TTM6 N6UKW6 A0A2J7QH94 E9IKF8
Pubmed
EMBL
AGBW02014789
OWR41062.1
KQ460883
KPJ11152.1
KQ459603
KPI92808.1
+ More
KZ149911 PZC78108.1 JTDY01004843 KOB67682.1 AK402758 BAM19368.1 ODYU01000745 SOQ35996.1 ODYU01006526 SOQ48459.1 BABH01003829 BABH01003830 BABH01003831 NWSH01001458 PCG71170.1 KQ460323 KPJ15970.1 KQ458756 KPJ05260.1 BABH01003834 AGBW02007651 OWR55012.1 KPJ05259.1 KPJ15971.1 ODYU01002091 SOQ39165.1 DS235784 EEB16960.1 HQ245150 AEB91339.1 HQ245152 AEB91341.1 GGMS01001005 MBY70208.1 GAPU01000071 JAB84774.1 CH477290 EAT44646.1 NEVH01005904 PNF38176.1 ABLF02037922 CH477801 EAT36071.1 GFXV01004112 MBW15917.1 GEDC01006623 JAS30675.1 CVRI01000019 CRK90469.1 AAZX01003729 GGMR01016673 MBY29292.1 AJVK01032026 JXUM01056073 JXUM01056074 KQ561895 KXJ77234.1 GL888284 EGI63349.1 GL763984 EFZ18940.1 ADTU01026685 JXUM01102122 JXUM01102123 JXUM01102124 JXUM01102125 KQ564702 KXJ71872.1 KQ980314 KYN16692.1 KQ971348 EFA05534.2 GAPU01000070 JAB84775.1 KQ981296 KYN43080.1 GL434548 EFN74792.1 GBYB01007495 JAG77262.1 GECU01009357 JAS98349.1 QOIP01000012 RLU15835.1 JTDY01001201 KOB74558.1 KK852802 KDR16287.1 EFZ18935.1 KQ459595 KPI96020.1 KPJ15961.1 GL436992 EFN71295.1 GL732535 EFX84005.1 KQ981281 KYN43299.1 ADTU01026680 GARF01000007 JAC88928.1 KQ759784 OAD62863.1 KQ976532 KYM81568.1 KQ979701 KYN19666.1 KQ982937 KYQ49100.1 ABLF02035756 ABLF02035762 KQ434869 KZC09328.1 KZC09314.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 KY618825 AQW43011.1 JXUM01024565 KQ560694 KXJ81242.1 LBMM01006355 KMQ90671.1 KK853018 KDR12457.1 GL888048 EGI68822.1 ABLF02037928 ACPB03007733 ACPB03007734 ACPB03007735 ACPB03007736 GEZM01076638 JAV63727.1 PCG71174.1 LBMM01006334 KMQ90686.1 GL448571 EFN84154.1 ATLV01012991 KE524833 KFB37100.1 JXUM01027342 KQ560787 KXJ80883.1 JXUM01024564 KXJ81241.1 NEVH01022362 PNF18879.1 AAAB01008844 EAA45200.5 ADMH02002030 ETN59864.1 GBYB01004178 JAG73945.1 GFXV01003241 MBW15046.1 DS232197 EDS37178.1 APCN01003125 AAZX01008029 AAZX01015187 GDIP01102390 JAM01325.1 GDIQ01125583 JAL26143.1 APGK01029196 KB740727 ENN79327.1 NEVH01013984 PNF27957.1 EFZ18947.1
KZ149911 PZC78108.1 JTDY01004843 KOB67682.1 AK402758 BAM19368.1 ODYU01000745 SOQ35996.1 ODYU01006526 SOQ48459.1 BABH01003829 BABH01003830 BABH01003831 NWSH01001458 PCG71170.1 KQ460323 KPJ15970.1 KQ458756 KPJ05260.1 BABH01003834 AGBW02007651 OWR55012.1 KPJ05259.1 KPJ15971.1 ODYU01002091 SOQ39165.1 DS235784 EEB16960.1 HQ245150 AEB91339.1 HQ245152 AEB91341.1 GGMS01001005 MBY70208.1 GAPU01000071 JAB84774.1 CH477290 EAT44646.1 NEVH01005904 PNF38176.1 ABLF02037922 CH477801 EAT36071.1 GFXV01004112 MBW15917.1 GEDC01006623 JAS30675.1 CVRI01000019 CRK90469.1 AAZX01003729 GGMR01016673 MBY29292.1 AJVK01032026 JXUM01056073 JXUM01056074 KQ561895 KXJ77234.1 GL888284 EGI63349.1 GL763984 EFZ18940.1 ADTU01026685 JXUM01102122 JXUM01102123 JXUM01102124 JXUM01102125 KQ564702 KXJ71872.1 KQ980314 KYN16692.1 KQ971348 EFA05534.2 GAPU01000070 JAB84775.1 KQ981296 KYN43080.1 GL434548 EFN74792.1 GBYB01007495 JAG77262.1 GECU01009357 JAS98349.1 QOIP01000012 RLU15835.1 JTDY01001201 KOB74558.1 KK852802 KDR16287.1 EFZ18935.1 KQ459595 KPI96020.1 KPJ15961.1 GL436992 EFN71295.1 GL732535 EFX84005.1 KQ981281 KYN43299.1 ADTU01026680 GARF01000007 JAC88928.1 KQ759784 OAD62863.1 KQ976532 KYM81568.1 KQ979701 KYN19666.1 KQ982937 KYQ49100.1 ABLF02035756 ABLF02035762 KQ434869 KZC09328.1 KZC09314.1 GEDC01024026 GEDC01011147 JAS13272.1 JAS26151.1 KY618825 AQW43011.1 JXUM01024565 KQ560694 KXJ81242.1 LBMM01006355 KMQ90671.1 KK853018 KDR12457.1 GL888048 EGI68822.1 ABLF02037928 ACPB03007733 ACPB03007734 ACPB03007735 ACPB03007736 GEZM01076638 JAV63727.1 PCG71174.1 LBMM01006334 KMQ90686.1 GL448571 EFN84154.1 ATLV01012991 KE524833 KFB37100.1 JXUM01027342 KQ560787 KXJ80883.1 JXUM01024564 KXJ81241.1 NEVH01022362 PNF18879.1 AAAB01008844 EAA45200.5 ADMH02002030 ETN59864.1 GBYB01004178 JAG73945.1 GFXV01003241 MBW15046.1 DS232197 EDS37178.1 APCN01003125 AAZX01008029 AAZX01015187 GDIP01102390 JAM01325.1 GDIQ01125583 JAL26143.1 APGK01029196 KB740727 ENN79327.1 NEVH01013984 PNF27957.1 EFZ18947.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000037510
UP000005204
UP000218220
+ More
UP000009046 UP000008820 UP000235965 UP000007819 UP000183832 UP000002358 UP000092462 UP000005203 UP000069940 UP000249989 UP000007755 UP000005205 UP000078492 UP000007266 UP000078541 UP000000311 UP000279307 UP000027135 UP000000305 UP000078540 UP000075809 UP000076502 UP000069272 UP000192223 UP000036403 UP000015103 UP000075882 UP000008237 UP000030765 UP000007062 UP000000673 UP000076407 UP000002320 UP000075840 UP000075880 UP000075902 UP000019118
UP000009046 UP000008820 UP000235965 UP000007819 UP000183832 UP000002358 UP000092462 UP000005203 UP000069940 UP000249989 UP000007755 UP000005205 UP000078492 UP000007266 UP000078541 UP000000311 UP000279307 UP000027135 UP000000305 UP000078540 UP000075809 UP000076502 UP000069272 UP000192223 UP000036403 UP000015103 UP000075882 UP000008237 UP000030765 UP000007062 UP000000673 UP000076407 UP000002320 UP000075840 UP000075880 UP000075902 UP000019118
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
A0A212EHV5
A0A194R0C6
A0A194PID4
A0A2W1BSS3
A0A0L7KWS0
I4DN78
+ More
A0A2H1V5E5 A0A2H1W5U4 H9JTZ4 H9JTZ5 A0A2A4JIB2 A0A194RF91 A0A194QK24 H9JTZ7 A0A212FMN5 A0A194QIN4 A0A194RJ52 A0A2H1VEA7 E0VUA4 F4ZNI1 F4ZNI3 A0A2S2PXF2 A0A1S4F6C0 W4VSN1 Q17DV5 A0A2J7RBG4 J9JXJ8 Q16P03 A0A1S4FU90 A0A2H8TP19 A0A1B6DYE3 A0A1J1HR16 K7J274 A0A2S2PIK7 A0A1B0D7W4 A0A088AJU4 A0A182GDJ6 F4WR74 E9IKG7 A0A158NUR3 A0A182H0W9 A0A151J323 D2A3N4 W4VS94 A0A151K0E9 E1ZVM3 A0A0C9RGK1 A0A1B6JGN2 A0A3L8D5U9 A0A0L7LG80 A0A067RC53 E9IKG5 A0A194PXZ1 A0A194RJ41 E2A5J5 E9G8N9 A0A195FTT2 A0A158NUU8 A0A0U9HSH0 A0A310SX81 A0A195BAK2 A0A195E3M0 A0A151WMQ2 J9JWV7 A0A154PDJ9 A0A182F7P4 A0A154PD41 A0A1B6DKE4 A0A2K8FTL4 A0A182H3F8 A0A1W4XEG1 A0A0J7KKC5 A0A067QS81 F4WAD9 J9JM93 T1HCA7 A0A1Y1KXV7 A0A182LAT9 A0A2A4JHW7 A0A0J7KKD8 E2BJJ7 A0A084VGK6 A0A182H5D1 A0A182H3F7 A0A2J7PRD5 Q7PFS6 W5JAQ1 A0A0C9PS40 A0A2H8TLM1 A0A182WV76 B0WYR8 A0A182I8V1 K7J283 A0A0P5W9T8 A0A0P5PUH8 A0A182JLA4 A0A182TTM6 N6UKW6 A0A2J7QH94 E9IKF8
A0A2H1V5E5 A0A2H1W5U4 H9JTZ4 H9JTZ5 A0A2A4JIB2 A0A194RF91 A0A194QK24 H9JTZ7 A0A212FMN5 A0A194QIN4 A0A194RJ52 A0A2H1VEA7 E0VUA4 F4ZNI1 F4ZNI3 A0A2S2PXF2 A0A1S4F6C0 W4VSN1 Q17DV5 A0A2J7RBG4 J9JXJ8 Q16P03 A0A1S4FU90 A0A2H8TP19 A0A1B6DYE3 A0A1J1HR16 K7J274 A0A2S2PIK7 A0A1B0D7W4 A0A088AJU4 A0A182GDJ6 F4WR74 E9IKG7 A0A158NUR3 A0A182H0W9 A0A151J323 D2A3N4 W4VS94 A0A151K0E9 E1ZVM3 A0A0C9RGK1 A0A1B6JGN2 A0A3L8D5U9 A0A0L7LG80 A0A067RC53 E9IKG5 A0A194PXZ1 A0A194RJ41 E2A5J5 E9G8N9 A0A195FTT2 A0A158NUU8 A0A0U9HSH0 A0A310SX81 A0A195BAK2 A0A195E3M0 A0A151WMQ2 J9JWV7 A0A154PDJ9 A0A182F7P4 A0A154PD41 A0A1B6DKE4 A0A2K8FTL4 A0A182H3F8 A0A1W4XEG1 A0A0J7KKC5 A0A067QS81 F4WAD9 J9JM93 T1HCA7 A0A1Y1KXV7 A0A182LAT9 A0A2A4JHW7 A0A0J7KKD8 E2BJJ7 A0A084VGK6 A0A182H5D1 A0A182H3F7 A0A2J7PRD5 Q7PFS6 W5JAQ1 A0A0C9PS40 A0A2H8TLM1 A0A182WV76 B0WYR8 A0A182I8V1 K7J283 A0A0P5W9T8 A0A0P5PUH8 A0A182JLA4 A0A182TTM6 N6UKW6 A0A2J7QH94 E9IKF8
PDB
5NCC
E-value=3.41962e-62,
Score=607
Ontologies
GO
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.633815
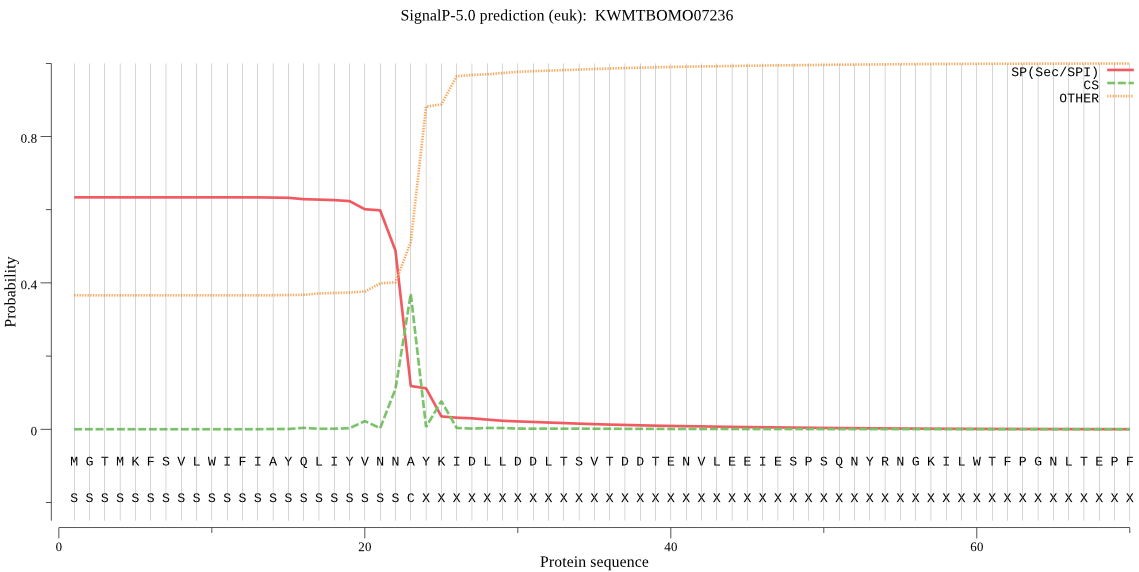
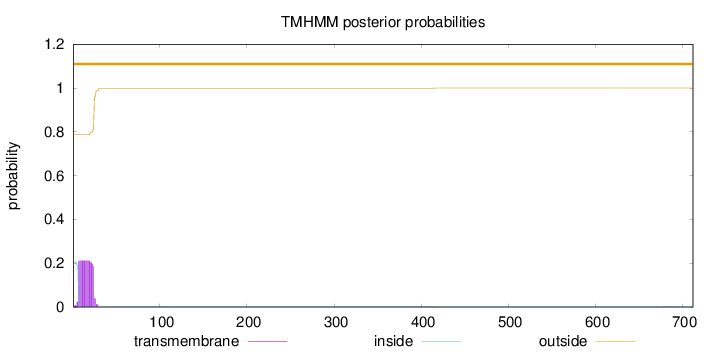
Length:
712
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.95182
Exp number, first 60 AAs:
3.93453
Total prob of N-in:
0.21213
outside
1 - 712
Population Genetic Test Statistics
Pi
23.336865
Theta
2.060307
Tajima's D
0.519397
CLR
0.528102
CSRT
0.575471226438678
Interpretation
Uncertain
