Gene
KWMTBOMO07233
Annotation
PREDICTED:_lipase_member_H-B-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.805
Sequence
CDS
ATGAACGCAGGTGTCTTCGCCTTCTTCTTTATCCTCGTCGGGAGCGGTCAATGGCAATTAATGTCATGTCATCGAGTACACTCGTCGGATTACTTCGATCCAAGCGCCTACATCGAAGATTGGACATTAGACAATGAAAATGAAACACTTCATTTGCCTTCGTCATACCTCCACAGCTTCGGATATAATCACTCGTGGAATGGTTACAGTAGCAATGAAGATGGCGCAAAATTGCGTTATTATTTCGGTTCAAGGAGCGATTATAAGGAATGGCCCTTGTCAGAAGCTCATAAAATTTTTTCATACAGTTGGTACGATCCCAGTAAGCCAACCGTGATTTTCACTCACGGTTTCACAGGTTTTCCCGAAGGTTTAGCGGTAGTTTCGGTAGTCGGTTTCTTCATAGAAGAAGGCAAGAGTAATGTGATACTTTTGAATTGGCAACATCTCGCTGCCCACGTGTATCCCAGCTTCGTGAATTCATACTTGAATTGGGCCGCACCAAACGCAATAAAGGTTGGAGCGGAGTTTGCGGAAACTCTGTTGAACTTGTCAAAGACGGGACTAGATTTGGGAAAGACCCACTTAGTTGGACATTCTCTTGGAGCGCATATCTTCGGGATTGCTGGCAATAGGATTTCGGAGAGAGGGGTGCAGTTACCTTGGATTACAGGATTGGATCCATCCAGTGCAGTGTTTGATATGAAACCTGCTCATATGAAACTGAACCCAAACTCTGCGCGGTACGTGTCAGTAGTACATTCTGATCCGAAAACCGTGGGCCAGTTTTACAACCTGGATGCCCAGCCGGTCGGTATGCTAGATTCTCCAGTGAAGATTTATGCAGTCACCATCGATGTTGGCAACTCCTAA
Protein
MNAGVFAFFFILVGSGQWQLMSCHRVHSSDYFDPSAYIEDWTLDNENETLHLPSSYLHSFGYNHSWNGYSSNEDGAKLRYYFGSRSDYKEWPLSEAHKIFSYSWYDPSKPTVIFTHGFTGFPEGLAVVSVVGFFIEEGKSNVILLNWQHLAAHVYPSFVNSYLNWAAPNAIKVGAEFAETLLNLSKTGLDLGKTHLVGHSLGAHIFGIAGNRISERGVQLPWITGLDPSSAVFDMKPAHMKLNPNSARYVSVVHSDPKTVGQFYNLDAQPVGMLDSPVKIYAVTIDVGNS
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
PDB
2PPL
E-value=1.95241e-13,
Score=182
Ontologies
PANTHER
Topology
Subcellular location
Secreted
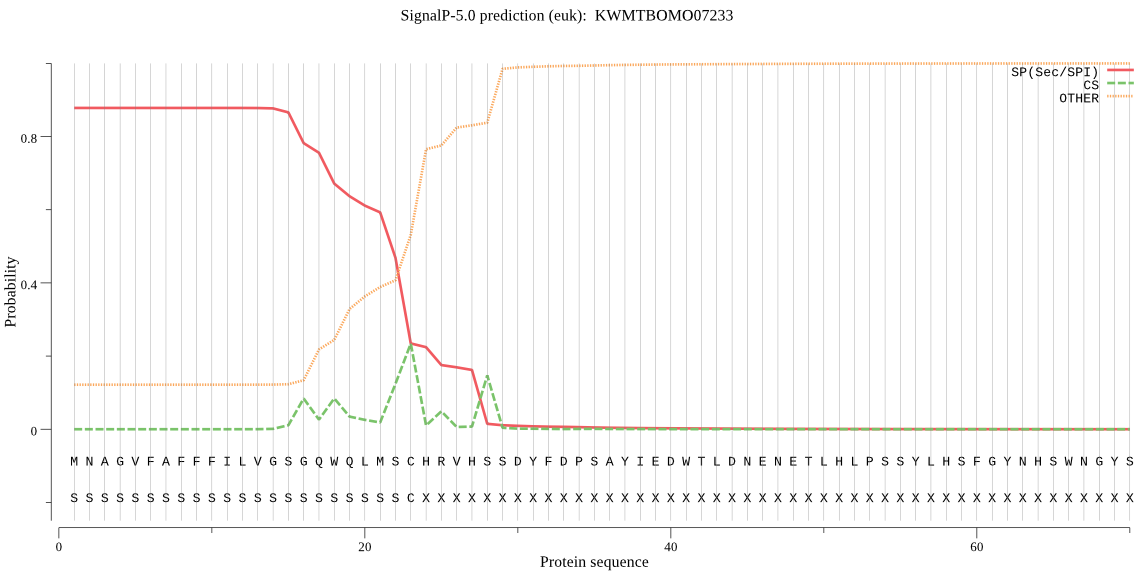
SignalP
Position: 1 - 23,
Likelihood: 0.878701
Length:
290
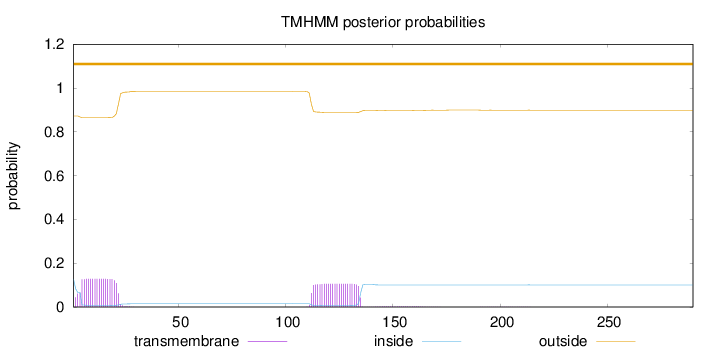
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.91295999999998
Exp number, first 60 AAs:
2.41434
Total prob of N-in:
0.12754
outside
1 - 290
Population Genetic Test Statistics
Pi
5.038703
Theta
14.693419
Tajima's D
-1.137753
CLR
1.421775
CSRT
0.109694515274236
Interpretation
Uncertain
