Gene
KWMTBOMO07229
Annotation
hypothetical_protein_KGM_12721_[Danaus_plexippus]
Full name
Palmitoyltransferase
Location in the cell
PlasmaMembrane Reliability : 4.547
Sequence
CDS
ATGAAGGCAATCGCAGATTTTGAACTAGCAACTGGTTCTTCGAAAGGCAAAGAATATAGCCCAGTACTAACTTCTGATTCATACTACTGCCAAGTATGCCAAATAGAAGTCAACGAAAGATTCTTTCATTCAATATGGTGGGACTGTTGTGTTTTGAGGCCTAATTACGTGTATTTCCTAGCTGGTCAGATATGTGCATTTGCAACACTTCTATTTGGAACCAACTTAGGCCTGACGACTGTGTGCCAACCATTCATTTTTTATGGATTAATATTATTGCCGGAGGATTGTAATGATGCATATTATGAGTTTGACTTATCACTCGGTTTTGTGACCTGTGTGTATGGTGTTGGATACCTTTTTGTTATTGCTTTAGTTCTAATAAGACAACTGCTCATCTATTTACCAAAATACAGTGAACCCCAGTGGAGAAAACTTGTGAACGTATTAAACGTGTGA
Protein
MKAIADFELATGSSKGKEYSPVLTSDSYYCQVCQIEVNERFFHSIWWDCCVLRPNYVYFLAGQICAFATLLFGTNLGLTTVCQPFIFYGLILLPEDCNDAYYEFDLSLGFVTCVYGVGYLFVIALVLIRQLLIYLPKYSEPQWRKLVNVLNV
Summary
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Similarity
Belongs to the DHHC palmitoyltransferase family.
Feature
chain Palmitoyltransferase
Uniprot
A0A212EHV7
S4PWZ2
A0A194PHK9
A0A2A4K901
A0A2Z5UJ38
A0A0N0PFK1
+ More
D6X334 A0A1Q3G3L0 A0A1B6DDN9 K7J1S5 A0A232F6Z0 A0A1B6C432 A0A1B6DGU8 A0A1B6DR47 A0A023EQ66 A0A182GQ16 J3JTP2 Q17BB7 A0A182W6R0 A0A1L8DY02 A0A1L8DY20 A0A2A3EJ45 A0A088AN39 A0A1Y9ITS8 A0A1L8DY56 A0A0M8ZYD0 Q7Q471 A0A1Y9J0D2 A0A182ICH8 A0A182L0Z5 A0A151JSC2 A0A182MI63 A0A1S4GYF7 A0A182V2Z2 A0A1L8DY51 A0A1L8DYU8 A0A182R2Z9 A0A182JY98 A0A182PCN6 A0A0L7RDY7 A0A195FTP3 A0A151X2W5 A0A154PQ99 V5GBN0 A0A1W4WW87 F4W5D4 A0A151IPK6 E2BT76 A0A158NYV3 A0A182UAU0 A0A182QSS4 A0A182JFF2 A0A182MYY4 E2ALD7 A0A084W4U3 A0A195BTJ3 A0A3B0JBY0 A0A3L8DXA4 A0A1B0CEG1 A0A182Y3H5 A0A182F128 A0A067RK59 B4I2U7 A0A0R3NZI4 X2J6Q6 B4JA63 Q9VQK9 A0A0J9QVJ4 B4Q919 B4G6Z1 B4NXC8 Q29M00 A0A023FBK5 B4KIL2 W5JFG1 A0A1A9XK03 B4MW18 A0A1A9ZWS3 A0A0M5J8U0 A0A1A9UQW8 A0A1W4UTQ5 A0A0P9BP43 A0A146L0D8 A0A146L0P6 A0A1B6M996 A0A0A9Y7X6 B3NAP8 J9JWL3 A0A1B0G2Z3 B3MLK3 A0A1B6F3D1
D6X334 A0A1Q3G3L0 A0A1B6DDN9 K7J1S5 A0A232F6Z0 A0A1B6C432 A0A1B6DGU8 A0A1B6DR47 A0A023EQ66 A0A182GQ16 J3JTP2 Q17BB7 A0A182W6R0 A0A1L8DY02 A0A1L8DY20 A0A2A3EJ45 A0A088AN39 A0A1Y9ITS8 A0A1L8DY56 A0A0M8ZYD0 Q7Q471 A0A1Y9J0D2 A0A182ICH8 A0A182L0Z5 A0A151JSC2 A0A182MI63 A0A1S4GYF7 A0A182V2Z2 A0A1L8DY51 A0A1L8DYU8 A0A182R2Z9 A0A182JY98 A0A182PCN6 A0A0L7RDY7 A0A195FTP3 A0A151X2W5 A0A154PQ99 V5GBN0 A0A1W4WW87 F4W5D4 A0A151IPK6 E2BT76 A0A158NYV3 A0A182UAU0 A0A182QSS4 A0A182JFF2 A0A182MYY4 E2ALD7 A0A084W4U3 A0A195BTJ3 A0A3B0JBY0 A0A3L8DXA4 A0A1B0CEG1 A0A182Y3H5 A0A182F128 A0A067RK59 B4I2U7 A0A0R3NZI4 X2J6Q6 B4JA63 Q9VQK9 A0A0J9QVJ4 B4Q919 B4G6Z1 B4NXC8 Q29M00 A0A023FBK5 B4KIL2 W5JFG1 A0A1A9XK03 B4MW18 A0A1A9ZWS3 A0A0M5J8U0 A0A1A9UQW8 A0A1W4UTQ5 A0A0P9BP43 A0A146L0D8 A0A146L0P6 A0A1B6M996 A0A0A9Y7X6 B3NAP8 J9JWL3 A0A1B0G2Z3 B3MLK3 A0A1B6F3D1
EC Number
2.3.1.225
Pubmed
22118469
23622113
26354079
18362917
19820115
20075255
+ More
28648823 24945155 26483478 22516182 17510324 12364791 14747013 17210077 20966253 21719571 20798317 21347285 24438588 30249741 25244985 24845553 17994087 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 25474469 20920257 23761445 26823975 25401762 18057021
28648823 24945155 26483478 22516182 17510324 12364791 14747013 17210077 20966253 21719571 20798317 21347285 24438588 30249741 25244985 24845553 17994087 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 25474469 20920257 23761445 26823975 25401762 18057021
EMBL
AGBW02014789
OWR41069.1
GAIX01005943
JAA86617.1
KQ459603
KPI92802.1
+ More
NWSH01000030 PCG80539.1 AP017527 BBB06902.1 LADJ01011234 KPJ20972.1 KQ971372 EFA10318.2 GFDL01000650 JAV34395.1 GEDC01013494 JAS23804.1 NNAY01000817 OXU26352.1 GEDC01029036 JAS08262.1 GEDC01012379 JAS24919.1 GEDC01009157 GEDC01003101 JAS28141.1 JAS34197.1 GAPW01001891 JAC11707.1 JXUM01079566 JXUM01079567 JXUM01079568 JXUM01079569 KQ563134 KXJ74427.1 BT126599 AEE61563.1 CH477324 EAT43529.2 GFDF01002744 JAV11340.1 GFDF01002743 JAV11341.1 KZ288245 PBC31209.1 GFDF01002745 JAV11339.1 KQ435824 KOX72089.1 AAAB01008964 EAA12334.4 APCN01000799 KQ978543 KYN30184.1 AXCM01001957 GFDF01002740 JAV11344.1 GFDF01002612 JAV11472.1 KQ414613 KOC69034.1 KQ981276 KYN43657.1 KQ982566 KYQ54787.1 KQ435037 KZC14095.1 GALX01000921 JAB67545.1 GL887645 EGI70584.1 KQ976837 KYN07961.1 GL450384 EFN81078.1 ADTU01004275 AXCN02000788 GL440604 EFN65742.1 ATLV01020383 KE525299 KFB45237.1 KQ976407 KYM91421.1 OUUW01000004 SPP79555.1 QOIP01000003 RLU25104.1 AJWK01008848 KK852424 KDR24177.1 CH480820 EDW54092.1 CH379060 KRT04307.1 AE014134 AHN54089.1 CH916368 EDW03737.1 AY069413 AAF51157.2 AAL39558.1 CM002910 KMY87809.1 CM000361 EDX03558.1 KMY87807.1 CH479180 EDW29190.1 CM000157 EDW87485.1 EAL33895.1 GBBI01000333 JAC18379.1 CH933807 EDW11355.1 ADMH02001645 ETN61620.1 CH963857 EDW75888.1 CP012523 ALC38606.1 CH902620 KPU73607.1 GDHC01017190 JAQ01439.1 GDHC01016468 JAQ02161.1 GEBQ01007465 JAT32512.1 GBHO01014447 GBHO01014446 GBHO01014444 JAG29157.1 JAG29158.1 JAG29160.1 CH954177 EDV57571.1 ABLF02031629 CCAG010003320 EDV31752.1 KPU73608.1 GECZ01025000 GECZ01016328 JAS44769.1 JAS53441.1
NWSH01000030 PCG80539.1 AP017527 BBB06902.1 LADJ01011234 KPJ20972.1 KQ971372 EFA10318.2 GFDL01000650 JAV34395.1 GEDC01013494 JAS23804.1 NNAY01000817 OXU26352.1 GEDC01029036 JAS08262.1 GEDC01012379 JAS24919.1 GEDC01009157 GEDC01003101 JAS28141.1 JAS34197.1 GAPW01001891 JAC11707.1 JXUM01079566 JXUM01079567 JXUM01079568 JXUM01079569 KQ563134 KXJ74427.1 BT126599 AEE61563.1 CH477324 EAT43529.2 GFDF01002744 JAV11340.1 GFDF01002743 JAV11341.1 KZ288245 PBC31209.1 GFDF01002745 JAV11339.1 KQ435824 KOX72089.1 AAAB01008964 EAA12334.4 APCN01000799 KQ978543 KYN30184.1 AXCM01001957 GFDF01002740 JAV11344.1 GFDF01002612 JAV11472.1 KQ414613 KOC69034.1 KQ981276 KYN43657.1 KQ982566 KYQ54787.1 KQ435037 KZC14095.1 GALX01000921 JAB67545.1 GL887645 EGI70584.1 KQ976837 KYN07961.1 GL450384 EFN81078.1 ADTU01004275 AXCN02000788 GL440604 EFN65742.1 ATLV01020383 KE525299 KFB45237.1 KQ976407 KYM91421.1 OUUW01000004 SPP79555.1 QOIP01000003 RLU25104.1 AJWK01008848 KK852424 KDR24177.1 CH480820 EDW54092.1 CH379060 KRT04307.1 AE014134 AHN54089.1 CH916368 EDW03737.1 AY069413 AAF51157.2 AAL39558.1 CM002910 KMY87809.1 CM000361 EDX03558.1 KMY87807.1 CH479180 EDW29190.1 CM000157 EDW87485.1 EAL33895.1 GBBI01000333 JAC18379.1 CH933807 EDW11355.1 ADMH02001645 ETN61620.1 CH963857 EDW75888.1 CP012523 ALC38606.1 CH902620 KPU73607.1 GDHC01017190 JAQ01439.1 GDHC01016468 JAQ02161.1 GEBQ01007465 JAT32512.1 GBHO01014447 GBHO01014446 GBHO01014444 JAG29157.1 JAG29158.1 JAG29160.1 CH954177 EDV57571.1 ABLF02031629 CCAG010003320 EDV31752.1 KPU73608.1 GECZ01025000 GECZ01016328 JAS44769.1 JAS53441.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000053240
UP000007266
UP000002358
+ More
UP000215335 UP000069940 UP000249989 UP000008820 UP000075920 UP000242457 UP000005203 UP000053105 UP000007062 UP000076407 UP000075840 UP000075882 UP000078492 UP000075883 UP000075903 UP000075900 UP000075881 UP000075885 UP000053825 UP000078541 UP000075809 UP000076502 UP000192223 UP000007755 UP000078542 UP000008237 UP000005205 UP000075902 UP000075886 UP000075880 UP000075884 UP000000311 UP000030765 UP000078540 UP000268350 UP000279307 UP000092461 UP000076408 UP000069272 UP000027135 UP000001292 UP000001819 UP000000803 UP000001070 UP000000304 UP000008744 UP000002282 UP000009192 UP000000673 UP000092443 UP000007798 UP000092445 UP000092553 UP000078200 UP000192221 UP000007801 UP000008711 UP000007819 UP000092444
UP000215335 UP000069940 UP000249989 UP000008820 UP000075920 UP000242457 UP000005203 UP000053105 UP000007062 UP000076407 UP000075840 UP000075882 UP000078492 UP000075883 UP000075903 UP000075900 UP000075881 UP000075885 UP000053825 UP000078541 UP000075809 UP000076502 UP000192223 UP000007755 UP000078542 UP000008237 UP000005205 UP000075902 UP000075886 UP000075880 UP000075884 UP000000311 UP000030765 UP000078540 UP000268350 UP000279307 UP000092461 UP000076408 UP000069272 UP000027135 UP000001292 UP000001819 UP000000803 UP000001070 UP000000304 UP000008744 UP000002282 UP000009192 UP000000673 UP000092443 UP000007798 UP000092445 UP000092553 UP000078200 UP000192221 UP000007801 UP000008711 UP000007819 UP000092444
Pfam
PF01529 DHHC
Interpro
ProteinModelPortal
A0A212EHV7
S4PWZ2
A0A194PHK9
A0A2A4K901
A0A2Z5UJ38
A0A0N0PFK1
+ More
D6X334 A0A1Q3G3L0 A0A1B6DDN9 K7J1S5 A0A232F6Z0 A0A1B6C432 A0A1B6DGU8 A0A1B6DR47 A0A023EQ66 A0A182GQ16 J3JTP2 Q17BB7 A0A182W6R0 A0A1L8DY02 A0A1L8DY20 A0A2A3EJ45 A0A088AN39 A0A1Y9ITS8 A0A1L8DY56 A0A0M8ZYD0 Q7Q471 A0A1Y9J0D2 A0A182ICH8 A0A182L0Z5 A0A151JSC2 A0A182MI63 A0A1S4GYF7 A0A182V2Z2 A0A1L8DY51 A0A1L8DYU8 A0A182R2Z9 A0A182JY98 A0A182PCN6 A0A0L7RDY7 A0A195FTP3 A0A151X2W5 A0A154PQ99 V5GBN0 A0A1W4WW87 F4W5D4 A0A151IPK6 E2BT76 A0A158NYV3 A0A182UAU0 A0A182QSS4 A0A182JFF2 A0A182MYY4 E2ALD7 A0A084W4U3 A0A195BTJ3 A0A3B0JBY0 A0A3L8DXA4 A0A1B0CEG1 A0A182Y3H5 A0A182F128 A0A067RK59 B4I2U7 A0A0R3NZI4 X2J6Q6 B4JA63 Q9VQK9 A0A0J9QVJ4 B4Q919 B4G6Z1 B4NXC8 Q29M00 A0A023FBK5 B4KIL2 W5JFG1 A0A1A9XK03 B4MW18 A0A1A9ZWS3 A0A0M5J8U0 A0A1A9UQW8 A0A1W4UTQ5 A0A0P9BP43 A0A146L0D8 A0A146L0P6 A0A1B6M996 A0A0A9Y7X6 B3NAP8 J9JWL3 A0A1B0G2Z3 B3MLK3 A0A1B6F3D1
D6X334 A0A1Q3G3L0 A0A1B6DDN9 K7J1S5 A0A232F6Z0 A0A1B6C432 A0A1B6DGU8 A0A1B6DR47 A0A023EQ66 A0A182GQ16 J3JTP2 Q17BB7 A0A182W6R0 A0A1L8DY02 A0A1L8DY20 A0A2A3EJ45 A0A088AN39 A0A1Y9ITS8 A0A1L8DY56 A0A0M8ZYD0 Q7Q471 A0A1Y9J0D2 A0A182ICH8 A0A182L0Z5 A0A151JSC2 A0A182MI63 A0A1S4GYF7 A0A182V2Z2 A0A1L8DY51 A0A1L8DYU8 A0A182R2Z9 A0A182JY98 A0A182PCN6 A0A0L7RDY7 A0A195FTP3 A0A151X2W5 A0A154PQ99 V5GBN0 A0A1W4WW87 F4W5D4 A0A151IPK6 E2BT76 A0A158NYV3 A0A182UAU0 A0A182QSS4 A0A182JFF2 A0A182MYY4 E2ALD7 A0A084W4U3 A0A195BTJ3 A0A3B0JBY0 A0A3L8DXA4 A0A1B0CEG1 A0A182Y3H5 A0A182F128 A0A067RK59 B4I2U7 A0A0R3NZI4 X2J6Q6 B4JA63 Q9VQK9 A0A0J9QVJ4 B4Q919 B4G6Z1 B4NXC8 Q29M00 A0A023FBK5 B4KIL2 W5JFG1 A0A1A9XK03 B4MW18 A0A1A9ZWS3 A0A0M5J8U0 A0A1A9UQW8 A0A1W4UTQ5 A0A0P9BP43 A0A146L0D8 A0A146L0P6 A0A1B6M996 A0A0A9Y7X6 B3NAP8 J9JWL3 A0A1B0G2Z3 B3MLK3 A0A1B6F3D1
Ontologies
GO
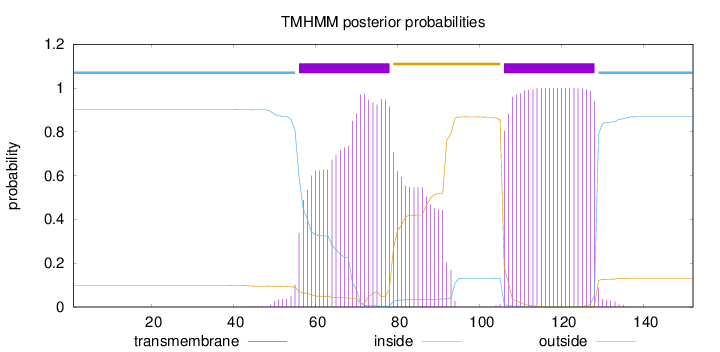
Topology
Length:
152
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
47.73339
Exp number, first 60 AAs:
2.9024
Total prob of N-in:
0.90131
inside
1 - 55
TMhelix
56 - 78
outside
79 - 105
TMhelix
106 - 128
inside
129 - 152
Population Genetic Test Statistics
Pi
30.647996
Theta
28.0068
Tajima's D
0.320216
CLR
2.296746
CSRT
0.462426878656067
Interpretation
Uncertain
