Gene
KWMTBOMO07228
Pre Gene Modal
BGIBMGA010579
Annotation
PREDICTED:_reticulocyte-binding_protein_2_homolog_a_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.952
Sequence
CDS
ATGAAATTTTTTTTGATCAATATTTATCAGAAGATTCTGTTAGAGAAGAAAAAATCTAATATTAATCAAGCAAAAGATTACACAAGTAGAGCAAAGTCTCAGGTTAAAGACTACAAAACTTATCAACAAAGGTACAAAGTTCAAAAACACATAATTGCTTTACAAAAGGCTAAACTTGAAGAACAGAATAGAATTATAGAAGAATTGAAATACAACAAAATTCTAGAAGCTTCGCGACAGTCAGTCGATGAAATGAGAGAAGAAGTTCGTAAATCTTATTTCGAAATAGATAGACAGTTAAAGCCTAAAATAAAATGTTTGACCAACGAACTAAAGATACCTGAAGTTGATGAATCGTCGTTGGTCCTTCATTGTTTAAAGGTGCCGCAGTTTTTACAAAGAATGGAGAAAAGAGCACGTGAAAGAGAAGAAAAACATGCTATGATACGAGAAAGACGTAAACAAATGGAAGAAGATAGAATTAGACTGAAGCAACAGGAAGAATTATCAAAGGCCGAAATGGATAAAGAAGAAAAACTTAGGAGGATTACAGAATTGCGTGAAAAAAGAAAAAAGGATAAAATAGAAAGCATTAGACGGAAGCAATACACTGAAAGAATGAGAGCTCTTAGAGTGATGGCTGACCTACACTACGAGAGATATTTAATGGTTAATTATGCTATCAGACCATTAAAAAAGCTATTAGAAATAAAACGATACAACGTAGAGATGGCGGAGTCACATTATAATTTCCAACTAAAGAAAAACGTATTCTTAAATTTCATGTGGTATACAGAAGATATGGTTTTCGAAAGAAATTATGTGGCACAAAATTTTTACCGAAAAAAAGTACTAAGAAAATACTTTGAAGGATTTAAGAAGAATCATTATGAATTCGTTTTGAAAATGCAAGTAGCCGTGGATTACTATGACTACTACGTTACTAATTTTATGTTTCGAAAATTTAAAGAAGGAATAAATGTTTTACGAAAGGAGTTTGAAATTAAAGTTCAAAGAGCTGATGCATACTATGAAAGTAATCTCCTGTTTAAGACTTTTACATGTTGGCGTACGTTGCCGGCTTTGAACGCACTCAAGAGAGAGCAAGAGGCGAGGAAAGCCAAATGGAGACAGAAGGTTTTACAAGTCGTCCCTGATTACACTCCGCCGGATGACTGA
Protein
MKFFLINIYQKILLEKKKSNINQAKDYTSRAKSQVKDYKTYQQRYKVQKHIIALQKAKLEEQNRIIEELKYNKILEASRQSVDEMREEVRKSYFEIDRQLKPKIKCLTNELKIPEVDESSLVLHCLKVPQFLQRMEKRAREREEKHAMIRERRKQMEEDRIRLKQQEELSKAEMDKEEKLRRITELREKRKKDKIESIRRKQYTERMRALRVMADLHYERYLMVNYAIRPLKKLLEIKRYNVEMAESHYNFQLKKNVFLNFMWYTEDMVFERNYVAQNFYRKKVLRKYFEGFKKNHYEFVLKMQVAVDYYDYYVTNFMFRKFKEGINVLRKEFEIKVQRADAYYESNLLFKTFTCWRTLPALNALKREQEARKAKWRQKVLQVVPDYTPPDD
Summary
Uniprot
H9JM27
A0A2A4K9Y8
A0A2H1VZC4
A0A0L7L8I4
A0A2Z5U853
A0A194PHB5
+ More
A0A194R033 A0A336MGR4 A0A336KYH0 A0A0L0CQY5 A0A1S3D6P4 A0A3Q0IY70 A0A1I8MY54 A0A1I8NSJ7 A0A3Q0J2L6 B4JA64 A0A067RK11 A0A3B0JB33 A0A1Q3FSW6 A0A182IYA5 A0A182H3Q4 Q29M01 B4G6Z0 A0A034VNW2 A0A1A9ZZP6 Q7PNE4 A0A182V2Z8 A0A182ICI0 A0A182UAC1 A0A182XL42 A0A1S4F9B6 A0A1S4GXS1 A0A182L0Z7 A0A182JY96 B4KIL1 A0A182R2Z7 A0A1A9XN16 A0A182PCN4 A0A182Y3H7 A0A0K8VI98 A0A182Q8R4 A0A0K8W2K8 B4NXC9 A0A182NU19 B0XJQ3 A0A1A9UZT4 A0A182W6R2 A0A084W4U1 A0A182MQ01 A0A0L7RDZ8 A0A1B0FR82 A0A1B0FRG5 W8BA84 A0A2A3EHL7 A0A088AN16 A0A182F130 A0A2P8YZ86 B3MLK2 B4LT20 A0A0Q9WQT4 B4MW19 A0A1W4V720 A0A0N0BEM7 B3NAP9 B4Q920 A0A0J9QUV9 Q32KC0 A0A1A9W6L5 Q9VQL0 M9PB20 M9PEB9 E2ALD8 D6WZU1 A0A2S2PHV8 A0A1B0CEG2 A0A151IPK1 A0A026W4N8 N6U2S8 A0A195FT73 A0A0J7NY44 A0A1J1IH99 A0A0M4EPR2 A0A0G2JUG8 A0A195BVB1 A0A151X2Y7 E9IYN2 A0A158NZB3 A0A232F7C5 F6SAD1 K7JL77 F4W5D3
A0A194R033 A0A336MGR4 A0A336KYH0 A0A0L0CQY5 A0A1S3D6P4 A0A3Q0IY70 A0A1I8MY54 A0A1I8NSJ7 A0A3Q0J2L6 B4JA64 A0A067RK11 A0A3B0JB33 A0A1Q3FSW6 A0A182IYA5 A0A182H3Q4 Q29M01 B4G6Z0 A0A034VNW2 A0A1A9ZZP6 Q7PNE4 A0A182V2Z8 A0A182ICI0 A0A182UAC1 A0A182XL42 A0A1S4F9B6 A0A1S4GXS1 A0A182L0Z7 A0A182JY96 B4KIL1 A0A182R2Z7 A0A1A9XN16 A0A182PCN4 A0A182Y3H7 A0A0K8VI98 A0A182Q8R4 A0A0K8W2K8 B4NXC9 A0A182NU19 B0XJQ3 A0A1A9UZT4 A0A182W6R2 A0A084W4U1 A0A182MQ01 A0A0L7RDZ8 A0A1B0FR82 A0A1B0FRG5 W8BA84 A0A2A3EHL7 A0A088AN16 A0A182F130 A0A2P8YZ86 B3MLK2 B4LT20 A0A0Q9WQT4 B4MW19 A0A1W4V720 A0A0N0BEM7 B3NAP9 B4Q920 A0A0J9QUV9 Q32KC0 A0A1A9W6L5 Q9VQL0 M9PB20 M9PEB9 E2ALD8 D6WZU1 A0A2S2PHV8 A0A1B0CEG2 A0A151IPK1 A0A026W4N8 N6U2S8 A0A195FT73 A0A0J7NY44 A0A1J1IH99 A0A0M4EPR2 A0A0G2JUG8 A0A195BVB1 A0A151X2Y7 E9IYN2 A0A158NZB3 A0A232F7C5 F6SAD1 K7JL77 F4W5D3
Pubmed
19121390
26227816
26354079
26108605
25315136
17994087
+ More
24845553 26483478 15632085 25348373 12364791 20966253 25244985 17550304 24438588 24495485 29403074 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317 18362917 19820115 24508170 23537049 15057822 21282665 21347285 28648823 19892987 20075255 21719571
24845553 26483478 15632085 25348373 12364791 20966253 25244985 17550304 24438588 24495485 29403074 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317 18362917 19820115 24508170 23537049 15057822 21282665 21347285 28648823 19892987 20075255 21719571
EMBL
BABH01005798
BABH01005799
NWSH01000030
PCG80540.1
ODYU01005376
SOQ46153.1
+ More
JTDY01002355 KOB71596.1 AP017527 BBB06901.1 KQ459603 KPI92801.1 KQ460883 KPJ11158.1 UFQT01001145 SSX29140.1 UFQS01001092 UFQT01001092 SSX08965.1 SSX28876.1 JRES01000145 KNC33839.1 CH916368 EDW03738.1 KK852424 KDR24176.1 OUUW01000004 SPP79554.1 GFDL01004345 JAV30700.1 JXUM01107781 JXUM01107782 KQ565173 KXJ71250.1 CH379060 EAL33894.2 CH479180 EDW29189.1 GAKP01015155 JAC43797.1 AAAB01008964 EAA12320.4 APCN01000799 CH933807 EDW11354.1 GDHF01014069 JAI38245.1 AXCN02000788 GDHF01007018 JAI45296.1 CM000157 EDW87486.2 DS233574 EDS30592.1 ATLV01020383 KE525299 KFB45235.1 AXCM01001956 KQ414613 KOC69035.1 CCAG010008016 GAMC01010958 GAMC01010956 JAB95597.1 KZ288245 PBC31210.1 PYGN01000273 PSN49566.1 CH902620 EDV31751.1 CH940649 EDW63851.1 CH963857 KRF98329.1 EDW75889.1 KQ435824 KOX72088.1 CH954177 EDV57572.2 CM000361 EDX03559.1 CM002910 KMY87811.1 KMY87812.1 BT023959 ABB36463.1 AE014134 AAF51156.2 AGB92518.1 AGB92517.1 GL440604 EFN65743.1 KQ971372 EFA09647.1 GGMR01016386 MBY29005.1 AJWK01008848 KQ976837 KYN07960.1 KK107419 EZA51045.1 APGK01041783 KB740997 ENN75875.1 KQ981276 KYN43658.1 LBMM01000872 KMQ97320.1 CVRI01000047 CRK98414.1 CP012523 ALC38607.1 AC114526 KQ976407 KYM91420.1 KQ982566 KYQ54786.1 GL767043 EFZ14276.1 ADTU01004274 NNAY01000817 OXU26348.1 GL887645 EGI70583.1
JTDY01002355 KOB71596.1 AP017527 BBB06901.1 KQ459603 KPI92801.1 KQ460883 KPJ11158.1 UFQT01001145 SSX29140.1 UFQS01001092 UFQT01001092 SSX08965.1 SSX28876.1 JRES01000145 KNC33839.1 CH916368 EDW03738.1 KK852424 KDR24176.1 OUUW01000004 SPP79554.1 GFDL01004345 JAV30700.1 JXUM01107781 JXUM01107782 KQ565173 KXJ71250.1 CH379060 EAL33894.2 CH479180 EDW29189.1 GAKP01015155 JAC43797.1 AAAB01008964 EAA12320.4 APCN01000799 CH933807 EDW11354.1 GDHF01014069 JAI38245.1 AXCN02000788 GDHF01007018 JAI45296.1 CM000157 EDW87486.2 DS233574 EDS30592.1 ATLV01020383 KE525299 KFB45235.1 AXCM01001956 KQ414613 KOC69035.1 CCAG010008016 GAMC01010958 GAMC01010956 JAB95597.1 KZ288245 PBC31210.1 PYGN01000273 PSN49566.1 CH902620 EDV31751.1 CH940649 EDW63851.1 CH963857 KRF98329.1 EDW75889.1 KQ435824 KOX72088.1 CH954177 EDV57572.2 CM000361 EDX03559.1 CM002910 KMY87811.1 KMY87812.1 BT023959 ABB36463.1 AE014134 AAF51156.2 AGB92518.1 AGB92517.1 GL440604 EFN65743.1 KQ971372 EFA09647.1 GGMR01016386 MBY29005.1 AJWK01008848 KQ976837 KYN07960.1 KK107419 EZA51045.1 APGK01041783 KB740997 ENN75875.1 KQ981276 KYN43658.1 LBMM01000872 KMQ97320.1 CVRI01000047 CRK98414.1 CP012523 ALC38607.1 AC114526 KQ976407 KYM91420.1 KQ982566 KYQ54786.1 GL767043 EFZ14276.1 ADTU01004274 NNAY01000817 OXU26348.1 GL887645 EGI70583.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000037069
+ More
UP000079169 UP000095301 UP000095300 UP000001070 UP000027135 UP000268350 UP000075880 UP000069940 UP000249989 UP000001819 UP000008744 UP000092445 UP000007062 UP000075903 UP000075840 UP000075902 UP000076407 UP000075882 UP000075881 UP000009192 UP000075900 UP000092443 UP000075885 UP000076408 UP000075886 UP000002282 UP000075884 UP000002320 UP000078200 UP000075920 UP000030765 UP000075883 UP000053825 UP000092444 UP000242457 UP000005203 UP000069272 UP000245037 UP000007801 UP000008792 UP000007798 UP000192221 UP000053105 UP000008711 UP000000304 UP000091820 UP000000803 UP000000311 UP000007266 UP000092461 UP000078542 UP000053097 UP000019118 UP000078541 UP000036403 UP000183832 UP000092553 UP000002494 UP000078540 UP000075809 UP000005205 UP000215335 UP000002281 UP000002358 UP000007755
UP000079169 UP000095301 UP000095300 UP000001070 UP000027135 UP000268350 UP000075880 UP000069940 UP000249989 UP000001819 UP000008744 UP000092445 UP000007062 UP000075903 UP000075840 UP000075902 UP000076407 UP000075882 UP000075881 UP000009192 UP000075900 UP000092443 UP000075885 UP000076408 UP000075886 UP000002282 UP000075884 UP000002320 UP000078200 UP000075920 UP000030765 UP000075883 UP000053825 UP000092444 UP000242457 UP000005203 UP000069272 UP000245037 UP000007801 UP000008792 UP000007798 UP000192221 UP000053105 UP000008711 UP000000304 UP000091820 UP000000803 UP000000311 UP000007266 UP000092461 UP000078542 UP000053097 UP000019118 UP000078541 UP000036403 UP000183832 UP000092553 UP000002494 UP000078540 UP000075809 UP000005205 UP000215335 UP000002281 UP000002358 UP000007755
Interpro
Gene 3D
ProteinModelPortal
H9JM27
A0A2A4K9Y8
A0A2H1VZC4
A0A0L7L8I4
A0A2Z5U853
A0A194PHB5
+ More
A0A194R033 A0A336MGR4 A0A336KYH0 A0A0L0CQY5 A0A1S3D6P4 A0A3Q0IY70 A0A1I8MY54 A0A1I8NSJ7 A0A3Q0J2L6 B4JA64 A0A067RK11 A0A3B0JB33 A0A1Q3FSW6 A0A182IYA5 A0A182H3Q4 Q29M01 B4G6Z0 A0A034VNW2 A0A1A9ZZP6 Q7PNE4 A0A182V2Z8 A0A182ICI0 A0A182UAC1 A0A182XL42 A0A1S4F9B6 A0A1S4GXS1 A0A182L0Z7 A0A182JY96 B4KIL1 A0A182R2Z7 A0A1A9XN16 A0A182PCN4 A0A182Y3H7 A0A0K8VI98 A0A182Q8R4 A0A0K8W2K8 B4NXC9 A0A182NU19 B0XJQ3 A0A1A9UZT4 A0A182W6R2 A0A084W4U1 A0A182MQ01 A0A0L7RDZ8 A0A1B0FR82 A0A1B0FRG5 W8BA84 A0A2A3EHL7 A0A088AN16 A0A182F130 A0A2P8YZ86 B3MLK2 B4LT20 A0A0Q9WQT4 B4MW19 A0A1W4V720 A0A0N0BEM7 B3NAP9 B4Q920 A0A0J9QUV9 Q32KC0 A0A1A9W6L5 Q9VQL0 M9PB20 M9PEB9 E2ALD8 D6WZU1 A0A2S2PHV8 A0A1B0CEG2 A0A151IPK1 A0A026W4N8 N6U2S8 A0A195FT73 A0A0J7NY44 A0A1J1IH99 A0A0M4EPR2 A0A0G2JUG8 A0A195BVB1 A0A151X2Y7 E9IYN2 A0A158NZB3 A0A232F7C5 F6SAD1 K7JL77 F4W5D3
A0A194R033 A0A336MGR4 A0A336KYH0 A0A0L0CQY5 A0A1S3D6P4 A0A3Q0IY70 A0A1I8MY54 A0A1I8NSJ7 A0A3Q0J2L6 B4JA64 A0A067RK11 A0A3B0JB33 A0A1Q3FSW6 A0A182IYA5 A0A182H3Q4 Q29M01 B4G6Z0 A0A034VNW2 A0A1A9ZZP6 Q7PNE4 A0A182V2Z8 A0A182ICI0 A0A182UAC1 A0A182XL42 A0A1S4F9B6 A0A1S4GXS1 A0A182L0Z7 A0A182JY96 B4KIL1 A0A182R2Z7 A0A1A9XN16 A0A182PCN4 A0A182Y3H7 A0A0K8VI98 A0A182Q8R4 A0A0K8W2K8 B4NXC9 A0A182NU19 B0XJQ3 A0A1A9UZT4 A0A182W6R2 A0A084W4U1 A0A182MQ01 A0A0L7RDZ8 A0A1B0FR82 A0A1B0FRG5 W8BA84 A0A2A3EHL7 A0A088AN16 A0A182F130 A0A2P8YZ86 B3MLK2 B4LT20 A0A0Q9WQT4 B4MW19 A0A1W4V720 A0A0N0BEM7 B3NAP9 B4Q920 A0A0J9QUV9 Q32KC0 A0A1A9W6L5 Q9VQL0 M9PB20 M9PEB9 E2ALD8 D6WZU1 A0A2S2PHV8 A0A1B0CEG2 A0A151IPK1 A0A026W4N8 N6U2S8 A0A195FT73 A0A0J7NY44 A0A1J1IH99 A0A0M4EPR2 A0A0G2JUG8 A0A195BVB1 A0A151X2Y7 E9IYN2 A0A158NZB3 A0A232F7C5 F6SAD1 K7JL77 F4W5D3
Ontologies
PANTHER
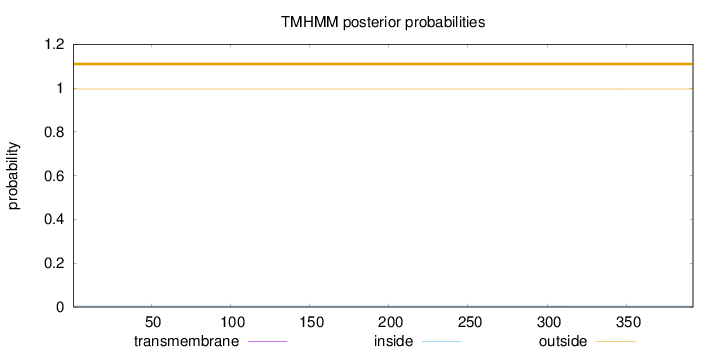
Topology
Length:
392
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00522
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00457
outside
1 - 392
Population Genetic Test Statistics
Pi
21.290832
Theta
17.522758
Tajima's D
0.595276
CLR
0.58582
CSRT
0.546972651367432
Interpretation
Uncertain
