Gene
KWMTBOMO07226 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010578
Annotation
PREDICTED:_L-2-hydroxyglutarate_dehydrogenase?_mitochondrial_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.946 Mitochondrial Reliability : 2.103
Sequence
CDS
ATGGCAATAGCGAAAAAATGCGTGCGATTTTTGCTAAATTTTAATTATCGTCATCCTGTTACCACTGGCACATTACGAAGATACAGTGCAGGTGCAGGTGATACTTCACAATTTGATGTAGCAGTGATTGGAGGAGGCATAGTAGGCTCTGCTTCAGCCAGGGAGTTAATATTAAAACACCCACATTTAAAGGTTGCTTTAGTTGAAAAAGAAAACCGATTTGCCTTCCACCAAAGTGGAAATAACAGTGGCGTGATTCATGCAGGTATTTATTATAAACCTGGTTCCCTTAAAGCAAAATTGTGTGTCAAAGGTCTGAACTTATCATATAAATATTTTGATCAGAAAGGAATCAAGTATTCCAAATGTGGTAAACTTATTGTGGCAACAGAGAGATCAGAAGTTCCTAGACTTTTTGATTTATATGAAAGGGGACTTAAGAATGGTGTTAAAGATATACAGCTCATGGATAGTAAAGAGATGAAAGAAATTGAACCACATTGCAAAGGATTACAGGCATTATGGTCTCCTCATACGGGTATTGTTAGTTGGGCTCAGGTTACAGACTCCTATGTTAAAGATTTTATTGAATGTGGTGGCAAAACCTATCTAAATTTTGAAGTAAAGAAATTTATAGAAGCTGAGTCTGCTGAGTATCCTGTAAAAATAATTGATACAGAAAATAATGTTATTAATGCAAAATATGTCCTCACATGCTGTGGACTGCACTCCGATAAAGTGGCAGTTTTAACAGGGTGTCCAGAAGAACCCAAAATAATACCATTTAGAGGTGAATACCTTTATTTGGTTCCAAACAAAAGTAAATTGACCAAAACCAATATATACCCTGTGCCTGACCCTAGGTTCCCATTTCTGGGTGTTCATTTAACACCAAGAATAGATGGCCGAGTTATTGTAGGACCAAATGCGATTTTAGCATTTTGCAAAGAAGGATATAGGTGGGGCGATTATAATATGCAGGAACTTAAAGAAATTATTGGTTTTTCCGGATTCCGCAAAATGGCTATGAAATATGCTAGTTTTGGTATCAAGGAAATGATAAGATCGGCTATAACACCTCTACAAGTTATGCAAGTTCGGAAGTATATACAGGAAATAACGTCAGCAGATGTTGAAAGGGGTCCAGCTGGTGTTAGGGCGCAAGCAATGGCTAAGGATGGTACATTAATAGAAGACTTTGTTTTTGATTCGAAACCTGGACATGGAGCAATAGGAAGTAGAGTACTCCATTGCCGAAATGCTCCATCCCCAGGAGCTACGTCATCACTTGCGATTGCTGAAATGGTTGTAGAGAAAATTGAAAAAGAATTTAAAATATGA
Protein
MAIAKKCVRFLLNFNYRHPVTTGTLRRYSAGAGDTSQFDVAVIGGGIVGSASARELILKHPHLKVALVEKENRFAFHQSGNNSGVIHAGIYYKPGSLKAKLCVKGLNLSYKYFDQKGIKYSKCGKLIVATERSEVPRLFDLYERGLKNGVKDIQLMDSKEMKEIEPHCKGLQALWSPHTGIVSWAQVTDSYVKDFIECGGKTYLNFEVKKFIEAESAEYPVKIIDTENNVINAKYVLTCCGLHSDKVAVLTGCPEEPKIIPFRGEYLYLVPNKSKLTKTNIYPVPDPRFPFLGVHLTPRIDGRVIVGPNAILAFCKEGYRWGDYNMQELKEIIGFSGFRKMAMKYASFGIKEMIRSAITPLQVMQVRKYIQEITSADVERGPAGVRAQAMAKDGTLIEDFVFDSKPGHGAIGSRVLHCRNAPSPGATSSLAIAEMVVEKIEKEFKI
Summary
Uniprot
H9JM26
A0A194R5M1
A0A194PJA1
A0A2A4K8K2
A0A212EHW3
A0A2H1VZC4
+ More
A0A1Q3G2J1 A0A0P6IW68 A0A182GIB6 A0A0P5XVS6 E9GS31 A0A0L0BRY1 A0A1I8N3Q7 A0A0N8BLN5 A0A0K8TMQ6 A0A0P5FZA2 A0A0P6GTV3 A0A0P6B2D8 A0A182YGZ7 T1PAR0 A0A182RBS9 A0A182JF95 A0A182WTE6 B4KJP5 A0A182V9J0 A0A084VP77 A0A182PMA7 A0A0P5YKP2 Q7Q3G9 A0A182HVE0 A0A182W9V8 A0A182TE56 A0A182M6X3 A0A182KG75 A0A1I8PXR2 A0A2M4BN32 A0A182Q1P6 A0A182NE98 A0A2M3Z9C7 A0A0P5Z792 A0A1I8PXN9 W5J6R2 A0A2M4BPD2 A0A2M3Z9G3 A0A2M3Z9E6 A0A182F4G6 A0A2J7RCV9 A0A0N8B994 A0A2M4AHR2 A0A2J7RCX2 A0A2J7RCW2 A0A0P4WQD0 A0A0M4E415 A0A0T6B500 A0A336LS65 A0A3B0K332 A0A1Y1LHF2 B4MZQ5 Q9VJ28 B4JBX8 A0A3B0JGN5 A0A0K8RJZ2 A0A0P5KJK5 B4I5N1 B4Q907 B3MPE7 B4LSJ3 A0A131YX23 A0A131Y0L2 B4PAD8 A0A1W4VXW0 A0A131Y1R4 A0A1B0C888 B4G7F2 A0A1A9Z0Q4 D3TP33 A0A1A9VXR6 A0A1B0G3T8 A0A1A9WK62 A0A034VDX0 A0A0K8UFZ2 E0VP15 Q29LU0 W8BE04 B3NL74 D2A384 A0A1Y1LMR5 A0A0P6GG25 A0A1W4WTY9 A0A1A9XTS0 A0A0P5C2H1 A0A1B0C0G8 A0A1D2N4I7 A0A1B6C7V0 A0A1E1XH88 A0A2B4RKF1 A0A1L8E053 A0A1B6MRH2 A0A1B6HDJ9 U4USS4
A0A1Q3G2J1 A0A0P6IW68 A0A182GIB6 A0A0P5XVS6 E9GS31 A0A0L0BRY1 A0A1I8N3Q7 A0A0N8BLN5 A0A0K8TMQ6 A0A0P5FZA2 A0A0P6GTV3 A0A0P6B2D8 A0A182YGZ7 T1PAR0 A0A182RBS9 A0A182JF95 A0A182WTE6 B4KJP5 A0A182V9J0 A0A084VP77 A0A182PMA7 A0A0P5YKP2 Q7Q3G9 A0A182HVE0 A0A182W9V8 A0A182TE56 A0A182M6X3 A0A182KG75 A0A1I8PXR2 A0A2M4BN32 A0A182Q1P6 A0A182NE98 A0A2M3Z9C7 A0A0P5Z792 A0A1I8PXN9 W5J6R2 A0A2M4BPD2 A0A2M3Z9G3 A0A2M3Z9E6 A0A182F4G6 A0A2J7RCV9 A0A0N8B994 A0A2M4AHR2 A0A2J7RCX2 A0A2J7RCW2 A0A0P4WQD0 A0A0M4E415 A0A0T6B500 A0A336LS65 A0A3B0K332 A0A1Y1LHF2 B4MZQ5 Q9VJ28 B4JBX8 A0A3B0JGN5 A0A0K8RJZ2 A0A0P5KJK5 B4I5N1 B4Q907 B3MPE7 B4LSJ3 A0A131YX23 A0A131Y0L2 B4PAD8 A0A1W4VXW0 A0A131Y1R4 A0A1B0C888 B4G7F2 A0A1A9Z0Q4 D3TP33 A0A1A9VXR6 A0A1B0G3T8 A0A1A9WK62 A0A034VDX0 A0A0K8UFZ2 E0VP15 Q29LU0 W8BE04 B3NL74 D2A384 A0A1Y1LMR5 A0A0P6GG25 A0A1W4WTY9 A0A1A9XTS0 A0A0P5C2H1 A0A1B0C0G8 A0A1D2N4I7 A0A1B6C7V0 A0A1E1XH88 A0A2B4RKF1 A0A1L8E053 A0A1B6MRH2 A0A1B6HDJ9 U4USS4
Pubmed
19121390
26354079
22118469
26999592
26483478
21292972
+ More
26108605 25315136 26369729 25244985 17994087 24438588 12364791 14747013 17210077 20920257 23761445 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26830274 17550304 20353571 25348373 20566863 15632085 24495485 18362917 19820115 27289101 28503490 23537049
26108605 25315136 26369729 25244985 17994087 24438588 12364791 14747013 17210077 20920257 23761445 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 26830274 17550304 20353571 25348373 20566863 15632085 24495485 18362917 19820115 27289101 28503490 23537049
EMBL
BABH01005795
KQ460883
KPJ11161.1
KQ459603
KPI92799.1
NWSH01000030
+ More
PCG80541.1 AGBW02014789 OWR41072.1 ODYU01005376 SOQ46153.1 GFDL01001008 JAV34037.1 GDUN01000716 JAN95203.1 JXUM01012104 JXUM01012105 JXUM01012106 KQ560345 KXJ82937.1 GDIQ01236133 GDIQ01167212 GDIP01066609 JAK15592.1 JAM37106.1 GL732561 EFX77633.1 JRES01001454 KNC22830.1 GDIQ01165579 JAK86146.1 GDAI01002398 JAI15205.1 GDIQ01253601 JAJ98123.1 GDIQ01028414 JAN66323.1 GDIP01020984 JAM82731.1 KA645240 AFP59869.1 CH933807 EDW11490.2 ATLV01014984 KE524999 KFB39771.1 GDIP01056453 JAM47262.1 AAAB01008964 EAA12921.2 APCN01002506 AXCM01000972 GGFJ01005349 MBW54490.1 AXCN02000864 GGFM01004372 MBW25123.1 GDIP01049390 JAM54325.1 ADMH02002133 ETN58459.1 GGFJ01005805 MBW54946.1 GGFM01004425 MBW25176.1 GGFM01004406 MBW25157.1 NEVH01005883 PNF38667.1 GDIQ01200307 JAK51418.1 GGFK01006966 MBW40287.1 PNF38668.1 PNF38665.1 GDRN01071243 JAI63730.1 CP012523 ALC38484.1 LJIG01009839 KRT82277.1 UFQT01000144 SSX20824.1 OUUW01000004 SPP79421.1 GEZM01057502 JAV72298.1 CH963920 EDW77840.1 AE014134 BT044190 AAF53729.1 ACH92255.1 AHN54590.1 AHN54591.1 CH916368 EDW04081.1 SPP79422.1 GADI01002949 JAA70859.1 GDIP01177098 GDIQ01184367 JAJ46304.1 JAK67358.1 CH480822 EDW55687.1 CM000361 CM002910 EDX05439.1 KMY90906.1 CH902620 EDV31243.1 CH940649 EDW64815.1 GEDV01005497 JAP83060.1 GEFM01002773 JAP73023.1 CM000158 EDW90346.1 GEFM01003358 JAP72438.1 AJWK01000274 CH479180 EDW29285.1 EZ423185 ADD19461.1 CCAG010005272 GAKP01018966 GAKP01018964 GAKP01018962 GAKP01018959 GAKP01018957 JAC39990.1 GDHF01026730 JAI25584.1 DS235354 EEB15121.1 CH379060 EAL33955.1 GAMC01011332 JAB95223.1 CH954179 EDV54724.1 KQ971338 EFA02276.1 GEZM01057503 JAV72297.1 GDIQ01035475 JAN59262.1 GDIP01177099 JAJ46303.1 JXJN01023591 LJIJ01000226 ODN00187.1 GEDC01027909 JAS09389.1 GFAC01000511 JAT98677.1 LSMT01000506 PFX16970.1 GFDF01002020 JAV12064.1 GEBQ01001437 JAT38540.1 GECU01034882 JAS72824.1 KB632433 ERL95573.1
PCG80541.1 AGBW02014789 OWR41072.1 ODYU01005376 SOQ46153.1 GFDL01001008 JAV34037.1 GDUN01000716 JAN95203.1 JXUM01012104 JXUM01012105 JXUM01012106 KQ560345 KXJ82937.1 GDIQ01236133 GDIQ01167212 GDIP01066609 JAK15592.1 JAM37106.1 GL732561 EFX77633.1 JRES01001454 KNC22830.1 GDIQ01165579 JAK86146.1 GDAI01002398 JAI15205.1 GDIQ01253601 JAJ98123.1 GDIQ01028414 JAN66323.1 GDIP01020984 JAM82731.1 KA645240 AFP59869.1 CH933807 EDW11490.2 ATLV01014984 KE524999 KFB39771.1 GDIP01056453 JAM47262.1 AAAB01008964 EAA12921.2 APCN01002506 AXCM01000972 GGFJ01005349 MBW54490.1 AXCN02000864 GGFM01004372 MBW25123.1 GDIP01049390 JAM54325.1 ADMH02002133 ETN58459.1 GGFJ01005805 MBW54946.1 GGFM01004425 MBW25176.1 GGFM01004406 MBW25157.1 NEVH01005883 PNF38667.1 GDIQ01200307 JAK51418.1 GGFK01006966 MBW40287.1 PNF38668.1 PNF38665.1 GDRN01071243 JAI63730.1 CP012523 ALC38484.1 LJIG01009839 KRT82277.1 UFQT01000144 SSX20824.1 OUUW01000004 SPP79421.1 GEZM01057502 JAV72298.1 CH963920 EDW77840.1 AE014134 BT044190 AAF53729.1 ACH92255.1 AHN54590.1 AHN54591.1 CH916368 EDW04081.1 SPP79422.1 GADI01002949 JAA70859.1 GDIP01177098 GDIQ01184367 JAJ46304.1 JAK67358.1 CH480822 EDW55687.1 CM000361 CM002910 EDX05439.1 KMY90906.1 CH902620 EDV31243.1 CH940649 EDW64815.1 GEDV01005497 JAP83060.1 GEFM01002773 JAP73023.1 CM000158 EDW90346.1 GEFM01003358 JAP72438.1 AJWK01000274 CH479180 EDW29285.1 EZ423185 ADD19461.1 CCAG010005272 GAKP01018966 GAKP01018964 GAKP01018962 GAKP01018959 GAKP01018957 JAC39990.1 GDHF01026730 JAI25584.1 DS235354 EEB15121.1 CH379060 EAL33955.1 GAMC01011332 JAB95223.1 CH954179 EDV54724.1 KQ971338 EFA02276.1 GEZM01057503 JAV72297.1 GDIQ01035475 JAN59262.1 GDIP01177099 JAJ46303.1 JXJN01023591 LJIJ01000226 ODN00187.1 GEDC01027909 JAS09389.1 GFAC01000511 JAT98677.1 LSMT01000506 PFX16970.1 GFDF01002020 JAV12064.1 GEBQ01001437 JAT38540.1 GECU01034882 JAS72824.1 KB632433 ERL95573.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000069940
+ More
UP000249989 UP000000305 UP000037069 UP000095301 UP000076408 UP000075900 UP000075880 UP000076407 UP000009192 UP000075903 UP000030765 UP000075885 UP000007062 UP000075840 UP000075920 UP000075902 UP000075883 UP000075881 UP000095300 UP000075886 UP000075884 UP000000673 UP000069272 UP000235965 UP000092553 UP000268350 UP000007798 UP000000803 UP000001070 UP000001292 UP000000304 UP000007801 UP000008792 UP000002282 UP000192221 UP000092461 UP000008744 UP000092445 UP000078200 UP000092444 UP000091820 UP000009046 UP000001819 UP000008711 UP000007266 UP000192223 UP000092443 UP000092460 UP000094527 UP000225706 UP000030742
UP000249989 UP000000305 UP000037069 UP000095301 UP000076408 UP000075900 UP000075880 UP000076407 UP000009192 UP000075903 UP000030765 UP000075885 UP000007062 UP000075840 UP000075920 UP000075902 UP000075883 UP000075881 UP000095300 UP000075886 UP000075884 UP000000673 UP000069272 UP000235965 UP000092553 UP000268350 UP000007798 UP000000803 UP000001070 UP000001292 UP000000304 UP000007801 UP000008792 UP000002282 UP000192221 UP000092461 UP000008744 UP000092445 UP000078200 UP000092444 UP000091820 UP000009046 UP000001819 UP000008711 UP000007266 UP000192223 UP000092443 UP000092460 UP000094527 UP000225706 UP000030742
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JM26
A0A194R5M1
A0A194PJA1
A0A2A4K8K2
A0A212EHW3
A0A2H1VZC4
+ More
A0A1Q3G2J1 A0A0P6IW68 A0A182GIB6 A0A0P5XVS6 E9GS31 A0A0L0BRY1 A0A1I8N3Q7 A0A0N8BLN5 A0A0K8TMQ6 A0A0P5FZA2 A0A0P6GTV3 A0A0P6B2D8 A0A182YGZ7 T1PAR0 A0A182RBS9 A0A182JF95 A0A182WTE6 B4KJP5 A0A182V9J0 A0A084VP77 A0A182PMA7 A0A0P5YKP2 Q7Q3G9 A0A182HVE0 A0A182W9V8 A0A182TE56 A0A182M6X3 A0A182KG75 A0A1I8PXR2 A0A2M4BN32 A0A182Q1P6 A0A182NE98 A0A2M3Z9C7 A0A0P5Z792 A0A1I8PXN9 W5J6R2 A0A2M4BPD2 A0A2M3Z9G3 A0A2M3Z9E6 A0A182F4G6 A0A2J7RCV9 A0A0N8B994 A0A2M4AHR2 A0A2J7RCX2 A0A2J7RCW2 A0A0P4WQD0 A0A0M4E415 A0A0T6B500 A0A336LS65 A0A3B0K332 A0A1Y1LHF2 B4MZQ5 Q9VJ28 B4JBX8 A0A3B0JGN5 A0A0K8RJZ2 A0A0P5KJK5 B4I5N1 B4Q907 B3MPE7 B4LSJ3 A0A131YX23 A0A131Y0L2 B4PAD8 A0A1W4VXW0 A0A131Y1R4 A0A1B0C888 B4G7F2 A0A1A9Z0Q4 D3TP33 A0A1A9VXR6 A0A1B0G3T8 A0A1A9WK62 A0A034VDX0 A0A0K8UFZ2 E0VP15 Q29LU0 W8BE04 B3NL74 D2A384 A0A1Y1LMR5 A0A0P6GG25 A0A1W4WTY9 A0A1A9XTS0 A0A0P5C2H1 A0A1B0C0G8 A0A1D2N4I7 A0A1B6C7V0 A0A1E1XH88 A0A2B4RKF1 A0A1L8E053 A0A1B6MRH2 A0A1B6HDJ9 U4USS4
A0A1Q3G2J1 A0A0P6IW68 A0A182GIB6 A0A0P5XVS6 E9GS31 A0A0L0BRY1 A0A1I8N3Q7 A0A0N8BLN5 A0A0K8TMQ6 A0A0P5FZA2 A0A0P6GTV3 A0A0P6B2D8 A0A182YGZ7 T1PAR0 A0A182RBS9 A0A182JF95 A0A182WTE6 B4KJP5 A0A182V9J0 A0A084VP77 A0A182PMA7 A0A0P5YKP2 Q7Q3G9 A0A182HVE0 A0A182W9V8 A0A182TE56 A0A182M6X3 A0A182KG75 A0A1I8PXR2 A0A2M4BN32 A0A182Q1P6 A0A182NE98 A0A2M3Z9C7 A0A0P5Z792 A0A1I8PXN9 W5J6R2 A0A2M4BPD2 A0A2M3Z9G3 A0A2M3Z9E6 A0A182F4G6 A0A2J7RCV9 A0A0N8B994 A0A2M4AHR2 A0A2J7RCX2 A0A2J7RCW2 A0A0P4WQD0 A0A0M4E415 A0A0T6B500 A0A336LS65 A0A3B0K332 A0A1Y1LHF2 B4MZQ5 Q9VJ28 B4JBX8 A0A3B0JGN5 A0A0K8RJZ2 A0A0P5KJK5 B4I5N1 B4Q907 B3MPE7 B4LSJ3 A0A131YX23 A0A131Y0L2 B4PAD8 A0A1W4VXW0 A0A131Y1R4 A0A1B0C888 B4G7F2 A0A1A9Z0Q4 D3TP33 A0A1A9VXR6 A0A1B0G3T8 A0A1A9WK62 A0A034VDX0 A0A0K8UFZ2 E0VP15 Q29LU0 W8BE04 B3NL74 D2A384 A0A1Y1LMR5 A0A0P6GG25 A0A1W4WTY9 A0A1A9XTS0 A0A0P5C2H1 A0A1B0C0G8 A0A1D2N4I7 A0A1B6C7V0 A0A1E1XH88 A0A2B4RKF1 A0A1L8E053 A0A1B6MRH2 A0A1B6HDJ9 U4USS4
PDB
3DME
E-value=1.59335e-26,
Score=297
Ontologies
PATHWAY
GO
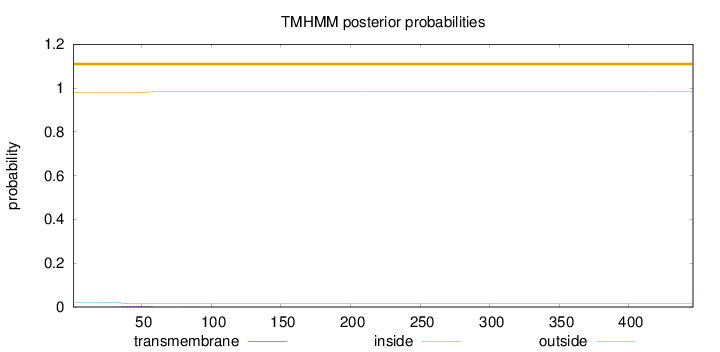
Topology
Length:
446
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0902099999999999
Exp number, first 60 AAs:
0.08555
Total prob of N-in:
0.01994
outside
1 - 446
Population Genetic Test Statistics
Pi
27.520501
Theta
32.210653
Tajima's D
0.063588
CLR
1.42409
CSRT
0.385330733463327
Interpretation
Uncertain
