Gene
KWMTBOMO07223
Pre Gene Modal
BGIBMGA010369
Annotation
PREDICTED:_aminoacylase-1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.065 PlasmaMembrane Reliability : 1.249
Sequence
CDS
ATGTTGAAATTTTTATGGTCTGTATTAATTATAATAGTCACGAGTGTGGAGACGGTTCCAGTGAAGAATCAATATGAATGTTATTTGAAAAAACCGGCAGTTCAAAGATTTAGGGAATACCTACAGATAGATACAAGTAAAGAAGAAAACTTAAAGTATGCCATCGAATTTTGGAAACGGCAAGCATTGGAAGCAAATCTTCCCTTTAATGTTTATAATCGACCTAATGGAAAACCTGTTGTCGTAATTACCGTTGAAGGAAGAGATCCGACGCTACCGGCCATCATGCTAAACTCTCACATGGACGTTGTGCCTGTTGAAGCGTCTGAATGGTCGCACCCGCCCTTCGCTGCAGTTATGGATGAAAATGGCGACATATTCGCGCGAGGAGCTCAGGATACAAAGGATGTTGGGATACAATATATGGAAGCTGTTAAGAGACTTATTCGTGATAACGTAACGTTGCTGCGCACTATATATCTTACGTTCATGCCCGATGAGGAAACTGGTGGGTACAAGGGCATGAAAATGTTTGTAGACACCGCAGAGTTTCAATCTTTAAACATCGGTTTCGCACTAGACGAGGGATTATCATCTACAGACGATACATTGTTTGCAACTTACACAGATCGGAGACCATGGCAAATCCAGTTTACTGTTCACGGTGTGGGTGGGCATGGTTATAGTATGCCAGAGAATACCGCAATGGAAAAGGTGTATAATTTAATTGACATAATAAGCAAGTATCGAGATACCCAAAGAAATTTATTGGAATCCTTAAATAGCCCCGATGAAGGAAGATACACCAGTGTCAATATAAACATAATTCGTGGTGGATACGCCACGAATGTTATACCAACGAATTTTACAATAACTATTGATATGAGGTTATCAACGGATGCAAAAATTTCGGATGTTCAAAATATGGTAAATACCTGGATGGAAAAAGCAGGCAATGATACTGAATTGAGTTTTATTCGGCGTGAAGACTTTTCAGGGGAGACTGATGTAGATGATTCAAATCCATTTTGGGTAGCTTTACGAACTGCTGTGACTAGTATGGGTTACAATGTCAAGCCGGTTGTTTGTTTGGCCACGTCTGATATGCTTGTGTTACGGAACAGAGGCGTACAAGCCCTCGGATTCGCCCCGAAGATCCGCACCGTTTCAAGACTTCACGCTAAAAACGAATACCAGAACGTCGTGGTATTTTTGAACGGCATCGATATCTATTACGAATAA
Protein
MLKFLWSVLIIIVTSVETVPVKNQYECYLKKPAVQRFREYLQIDTSKEENLKYAIEFWKRQALEANLPFNVYNRPNGKPVVVITVEGRDPTLPAIMLNSHMDVVPVEASEWSHPPFAAVMDENGDIFARGAQDTKDVGIQYMEAVKRLIRDNVTLLRTIYLTFMPDEETGGYKGMKMFVDTAEFQSLNIGFALDEGLSSTDDTLFATYTDRRPWQIQFTVHGVGGHGYSMPENTAMEKVYNLIDIISKYRDTQRNLLESLNSPDEGRYTSVNINIIRGGYATNVIPTNFTITIDMRLSTDAKISDVQNMVNTWMEKAGNDTELSFIRREDFSGETDVDDSNPFWVALRTAVTSMGYNVKPVVCLATSDMLVLRNRGVQALGFAPKIRTVSRLHAKNEYQNVVVFLNGIDIYYE
Summary
Uniprot
A0A2A4K8E3
A0A2H1VW17
A0A194R037
A0A194PHB0
A0A3S2P8Q0
A0A2J7RHR6
+ More
J9K9W5 C4WWX8 A0A1S3I4V8 A0A0L7RA17 A0A067R0Q5 A0A1Y1LZ21 A0A0A1XTF9 A0A2S2PKJ6 E2AMU9 W8ASK2 A0A310SSE3 A0A1L8E2D6 B4PN44 B0X290 A0A1L8E2H7 U5EYC3 A0A1B0CBE6 A0A034WAS3 B4K4M7 T1P9J4 E2BIK0 A0A2H8TPN6 A0A2S2QCL6 E2BIJ9 A0A1W4UR39 B4R1D2 B3LV52 V5G640 A0A2P8Y3J1 A0A151JMI0 A0A2H8TZZ1 A0A1Y1NL69 A0A0L0C800 Q9VCQ9 U4U9N9 A0A2A3EEL9 B3LV53 D6X327 A0A194PTR5 B3LVY1 E0VB00 B4NJN7 A0A0A1WGR0 A0A087ZSU7 Q9VCQ8 B4HFB1 A0A2T7NUK8 A0A182GM33 B4JDK8 A0A0M4E350 E9IDS2 Q8MRB5 B4K1D6 B3LV51 B4R1D3 W8BF16 F7CW09 R7TAZ0 A0A195FC31 A0A139WBF2 B0X291 I3M0R1 A0A1B6DJD3 A0A1Y1MM93 A0A069DSV9 A0A1W4VMJ7 T1HYA1 A0A1B0CLK7 A0A1Q3FN65 B4GP72 Q29BN1 A0A212FLD6 A0A158NK49 A0A1B6MHW6 W8BVI0 A0A1W4UQK5 A0A0L8GC35 A0A1B6M304 A0A0M8ZT35 A0A0M5J5K6 A0A1W4V4I1 B0WSY8 A0A182XY72 B4KH74 B4PN42 A0A0K8WFI1 Q9VCR0 A0A154PAG0 G3VXW2 Q16QR1 B4R1D1 A0A2Y9QU89 B4HFB0 W5JWJ1
J9K9W5 C4WWX8 A0A1S3I4V8 A0A0L7RA17 A0A067R0Q5 A0A1Y1LZ21 A0A0A1XTF9 A0A2S2PKJ6 E2AMU9 W8ASK2 A0A310SSE3 A0A1L8E2D6 B4PN44 B0X290 A0A1L8E2H7 U5EYC3 A0A1B0CBE6 A0A034WAS3 B4K4M7 T1P9J4 E2BIK0 A0A2H8TPN6 A0A2S2QCL6 E2BIJ9 A0A1W4UR39 B4R1D2 B3LV52 V5G640 A0A2P8Y3J1 A0A151JMI0 A0A2H8TZZ1 A0A1Y1NL69 A0A0L0C800 Q9VCQ9 U4U9N9 A0A2A3EEL9 B3LV53 D6X327 A0A194PTR5 B3LVY1 E0VB00 B4NJN7 A0A0A1WGR0 A0A087ZSU7 Q9VCQ8 B4HFB1 A0A2T7NUK8 A0A182GM33 B4JDK8 A0A0M4E350 E9IDS2 Q8MRB5 B4K1D6 B3LV51 B4R1D3 W8BF16 F7CW09 R7TAZ0 A0A195FC31 A0A139WBF2 B0X291 I3M0R1 A0A1B6DJD3 A0A1Y1MM93 A0A069DSV9 A0A1W4VMJ7 T1HYA1 A0A1B0CLK7 A0A1Q3FN65 B4GP72 Q29BN1 A0A212FLD6 A0A158NK49 A0A1B6MHW6 W8BVI0 A0A1W4UQK5 A0A0L8GC35 A0A1B6M304 A0A0M8ZT35 A0A0M5J5K6 A0A1W4V4I1 B0WSY8 A0A182XY72 B4KH74 B4PN42 A0A0K8WFI1 Q9VCR0 A0A154PAG0 G3VXW2 Q16QR1 B4R1D1 A0A2Y9QU89 B4HFB0 W5JWJ1
Pubmed
26354079
26383154
24845553
28004739
25830018
20798317
+ More
24495485 17994087 17550304 25348373 25315136 29403074 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 18362917 19820115 20566863 26483478 21282665 17495919 23254933 26334808 15632085 22118469 21347285 25244985 21709235 17510324 20920257 23761445
24495485 17994087 17550304 25348373 25315136 29403074 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 18362917 19820115 20566863 26483478 21282665 17495919 23254933 26334808 15632085 22118469 21347285 25244985 21709235 17510324 20920257 23761445
EMBL
NWSH01000030
PCG80545.1
ODYU01004628
SOQ44702.1
KQ460883
KPJ11163.1
+ More
KQ459603 KPI92796.1 RSAL01000001 RVE55108.1 NEVH01003737 PNF40375.1 ABLF02019043 AK342336 BAH72398.1 KQ414619 KOC67717.1 KK852801 KDR16327.1 GEZM01044285 GEZM01044282 JAV78331.1 GBXI01000304 JAD13988.1 GGMR01017295 MBY29914.1 GL440936 EFN65223.1 GAMC01014710 JAB91845.1 KQ760974 OAD58563.1 GFDF01001181 JAV12903.1 CM000160 EDW98099.1 DS232283 EDS39113.1 GFDF01001166 JAV12918.1 GANO01002088 JAB57783.1 AJWK01005087 GAKP01007163 JAC51789.1 CH933806 EDW15003.1 KA644835 AFP59464.1 GL448516 EFN84447.1 GFXV01004155 MBW15960.1 GGMS01006178 MBY75381.1 EFN84446.1 CM000364 EDX14018.1 CH902617 EDV42524.1 GALX01002936 JAB65530.1 PYGN01000970 PSN38838.1 KQ978928 KYN27485.1 GFXV01007267 MBW19072.1 GEZM01003849 JAV96527.1 JRES01000781 KNC28370.1 AE014297 BT023493 AAF56097.1 AAY84893.1 KB631843 ERL86650.1 KZ288280 PBC29616.1 EDV42525.1 KQ971372 EFA10313.1 KQ459592 KPI96806.1 EDV41514.2 DS235019 EEB10556.1 CH964272 EDW84999.1 GBXI01016451 GBXI01012706 GBXI01003387 JAC97840.1 JAD01586.1 JAD10905.1 BT050439 AAF56098.2 ACJ13146.1 CH480815 EDW43289.1 PZQS01000009 PVD24842.1 JXUM01073756 JXUM01073757 JXUM01073758 JXUM01073759 KQ562794 KXJ75120.1 CH916368 EDW03378.1 CP012523 ALC40178.1 GL762535 EFZ21239.1 AY121680 AAM52007.1 CH917093 EDW04843.1 EDV42523.1 EDX14019.1 GAMC01014709 JAB91846.1 AMQN01014106 KB310746 ELT90864.1 KQ981693 KYN37609.1 KYB25257.1 EDS39114.1 AGTP01046271 GEDC01011484 JAS25814.1 GEZM01027254 JAV86791.1 GBGD01001711 JAC87178.1 ACPB03023997 ACPB03025303 AJWK01017404 AJWK01017405 GFDL01005996 JAV29049.1 CH479186 EDW38955.1 CM000070 EAL26967.2 AGBW02007778 OWR54568.1 ADTU01018659 ADTU01018660 GEBQ01004471 JAT35506.1 GAMC01003353 JAC03203.1 KQ422866 KOF74085.1 GEBQ01009662 JAT30315.1 KQ435903 KOX69073.1 CP012526 ALC47906.1 DS232078 EDS34121.1 CH933807 EDW13291.1 EDW98097.1 GDHF01002535 JAI49779.1 AAF56096.1 AGB96239.1 KQ434857 KZC08889.1 AEFK01086815 AEFK01086816 AEFK01086817 CH477736 EAT36741.1 EDX14017.1 EDW43288.1 ADMH02000148 ETN67605.1
KQ459603 KPI92796.1 RSAL01000001 RVE55108.1 NEVH01003737 PNF40375.1 ABLF02019043 AK342336 BAH72398.1 KQ414619 KOC67717.1 KK852801 KDR16327.1 GEZM01044285 GEZM01044282 JAV78331.1 GBXI01000304 JAD13988.1 GGMR01017295 MBY29914.1 GL440936 EFN65223.1 GAMC01014710 JAB91845.1 KQ760974 OAD58563.1 GFDF01001181 JAV12903.1 CM000160 EDW98099.1 DS232283 EDS39113.1 GFDF01001166 JAV12918.1 GANO01002088 JAB57783.1 AJWK01005087 GAKP01007163 JAC51789.1 CH933806 EDW15003.1 KA644835 AFP59464.1 GL448516 EFN84447.1 GFXV01004155 MBW15960.1 GGMS01006178 MBY75381.1 EFN84446.1 CM000364 EDX14018.1 CH902617 EDV42524.1 GALX01002936 JAB65530.1 PYGN01000970 PSN38838.1 KQ978928 KYN27485.1 GFXV01007267 MBW19072.1 GEZM01003849 JAV96527.1 JRES01000781 KNC28370.1 AE014297 BT023493 AAF56097.1 AAY84893.1 KB631843 ERL86650.1 KZ288280 PBC29616.1 EDV42525.1 KQ971372 EFA10313.1 KQ459592 KPI96806.1 EDV41514.2 DS235019 EEB10556.1 CH964272 EDW84999.1 GBXI01016451 GBXI01012706 GBXI01003387 JAC97840.1 JAD01586.1 JAD10905.1 BT050439 AAF56098.2 ACJ13146.1 CH480815 EDW43289.1 PZQS01000009 PVD24842.1 JXUM01073756 JXUM01073757 JXUM01073758 JXUM01073759 KQ562794 KXJ75120.1 CH916368 EDW03378.1 CP012523 ALC40178.1 GL762535 EFZ21239.1 AY121680 AAM52007.1 CH917093 EDW04843.1 EDV42523.1 EDX14019.1 GAMC01014709 JAB91846.1 AMQN01014106 KB310746 ELT90864.1 KQ981693 KYN37609.1 KYB25257.1 EDS39114.1 AGTP01046271 GEDC01011484 JAS25814.1 GEZM01027254 JAV86791.1 GBGD01001711 JAC87178.1 ACPB03023997 ACPB03025303 AJWK01017404 AJWK01017405 GFDL01005996 JAV29049.1 CH479186 EDW38955.1 CM000070 EAL26967.2 AGBW02007778 OWR54568.1 ADTU01018659 ADTU01018660 GEBQ01004471 JAT35506.1 GAMC01003353 JAC03203.1 KQ422866 KOF74085.1 GEBQ01009662 JAT30315.1 KQ435903 KOX69073.1 CP012526 ALC47906.1 DS232078 EDS34121.1 CH933807 EDW13291.1 EDW98097.1 GDHF01002535 JAI49779.1 AAF56096.1 AGB96239.1 KQ434857 KZC08889.1 AEFK01086815 AEFK01086816 AEFK01086817 CH477736 EAT36741.1 EDX14017.1 EDW43288.1 ADMH02000148 ETN67605.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000283053
UP000235965
UP000007819
+ More
UP000085678 UP000053825 UP000027135 UP000000311 UP000002282 UP000002320 UP000092461 UP000009192 UP000095301 UP000008237 UP000192221 UP000000304 UP000007801 UP000245037 UP000078492 UP000037069 UP000000803 UP000030742 UP000242457 UP000007266 UP000009046 UP000007798 UP000005203 UP000001292 UP000245119 UP000069940 UP000249989 UP000001070 UP000092553 UP000002280 UP000014760 UP000078541 UP000005215 UP000015103 UP000008744 UP000001819 UP000007151 UP000005205 UP000053454 UP000053105 UP000076408 UP000076502 UP000007648 UP000008820 UP000248480 UP000000673
UP000085678 UP000053825 UP000027135 UP000000311 UP000002282 UP000002320 UP000092461 UP000009192 UP000095301 UP000008237 UP000192221 UP000000304 UP000007801 UP000245037 UP000078492 UP000037069 UP000000803 UP000030742 UP000242457 UP000007266 UP000009046 UP000007798 UP000005203 UP000001292 UP000245119 UP000069940 UP000249989 UP000001070 UP000092553 UP000002280 UP000014760 UP000078541 UP000005215 UP000015103 UP000008744 UP000001819 UP000007151 UP000005205 UP000053454 UP000053105 UP000076408 UP000076502 UP000007648 UP000008820 UP000248480 UP000000673
PRIDE
Interpro
SUPFAM
SSF55031
SSF55031
ProteinModelPortal
A0A2A4K8E3
A0A2H1VW17
A0A194R037
A0A194PHB0
A0A3S2P8Q0
A0A2J7RHR6
+ More
J9K9W5 C4WWX8 A0A1S3I4V8 A0A0L7RA17 A0A067R0Q5 A0A1Y1LZ21 A0A0A1XTF9 A0A2S2PKJ6 E2AMU9 W8ASK2 A0A310SSE3 A0A1L8E2D6 B4PN44 B0X290 A0A1L8E2H7 U5EYC3 A0A1B0CBE6 A0A034WAS3 B4K4M7 T1P9J4 E2BIK0 A0A2H8TPN6 A0A2S2QCL6 E2BIJ9 A0A1W4UR39 B4R1D2 B3LV52 V5G640 A0A2P8Y3J1 A0A151JMI0 A0A2H8TZZ1 A0A1Y1NL69 A0A0L0C800 Q9VCQ9 U4U9N9 A0A2A3EEL9 B3LV53 D6X327 A0A194PTR5 B3LVY1 E0VB00 B4NJN7 A0A0A1WGR0 A0A087ZSU7 Q9VCQ8 B4HFB1 A0A2T7NUK8 A0A182GM33 B4JDK8 A0A0M4E350 E9IDS2 Q8MRB5 B4K1D6 B3LV51 B4R1D3 W8BF16 F7CW09 R7TAZ0 A0A195FC31 A0A139WBF2 B0X291 I3M0R1 A0A1B6DJD3 A0A1Y1MM93 A0A069DSV9 A0A1W4VMJ7 T1HYA1 A0A1B0CLK7 A0A1Q3FN65 B4GP72 Q29BN1 A0A212FLD6 A0A158NK49 A0A1B6MHW6 W8BVI0 A0A1W4UQK5 A0A0L8GC35 A0A1B6M304 A0A0M8ZT35 A0A0M5J5K6 A0A1W4V4I1 B0WSY8 A0A182XY72 B4KH74 B4PN42 A0A0K8WFI1 Q9VCR0 A0A154PAG0 G3VXW2 Q16QR1 B4R1D1 A0A2Y9QU89 B4HFB0 W5JWJ1
J9K9W5 C4WWX8 A0A1S3I4V8 A0A0L7RA17 A0A067R0Q5 A0A1Y1LZ21 A0A0A1XTF9 A0A2S2PKJ6 E2AMU9 W8ASK2 A0A310SSE3 A0A1L8E2D6 B4PN44 B0X290 A0A1L8E2H7 U5EYC3 A0A1B0CBE6 A0A034WAS3 B4K4M7 T1P9J4 E2BIK0 A0A2H8TPN6 A0A2S2QCL6 E2BIJ9 A0A1W4UR39 B4R1D2 B3LV52 V5G640 A0A2P8Y3J1 A0A151JMI0 A0A2H8TZZ1 A0A1Y1NL69 A0A0L0C800 Q9VCQ9 U4U9N9 A0A2A3EEL9 B3LV53 D6X327 A0A194PTR5 B3LVY1 E0VB00 B4NJN7 A0A0A1WGR0 A0A087ZSU7 Q9VCQ8 B4HFB1 A0A2T7NUK8 A0A182GM33 B4JDK8 A0A0M4E350 E9IDS2 Q8MRB5 B4K1D6 B3LV51 B4R1D3 W8BF16 F7CW09 R7TAZ0 A0A195FC31 A0A139WBF2 B0X291 I3M0R1 A0A1B6DJD3 A0A1Y1MM93 A0A069DSV9 A0A1W4VMJ7 T1HYA1 A0A1B0CLK7 A0A1Q3FN65 B4GP72 Q29BN1 A0A212FLD6 A0A158NK49 A0A1B6MHW6 W8BVI0 A0A1W4UQK5 A0A0L8GC35 A0A1B6M304 A0A0M8ZT35 A0A0M5J5K6 A0A1W4V4I1 B0WSY8 A0A182XY72 B4KH74 B4PN42 A0A0K8WFI1 Q9VCR0 A0A154PAG0 G3VXW2 Q16QR1 B4R1D1 A0A2Y9QU89 B4HFB0 W5JWJ1
PDB
1Q7L
E-value=9.03827e-42,
Score=428
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.989491
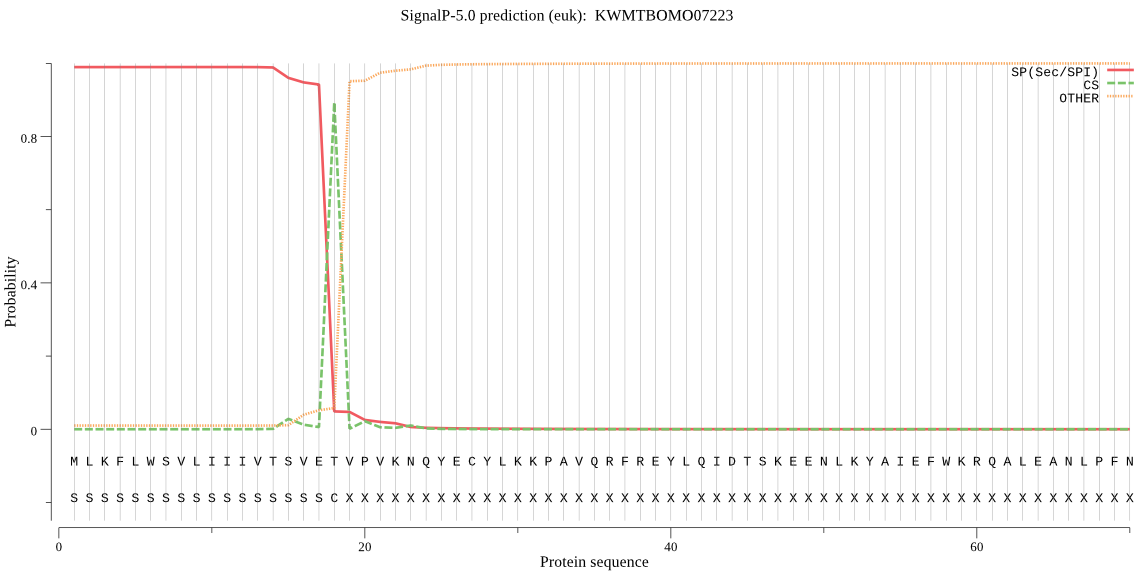
Length:
413
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.302430000000001
Exp number, first 60 AAs:
0.28565
Total prob of N-in:
0.01167
outside
1 - 413
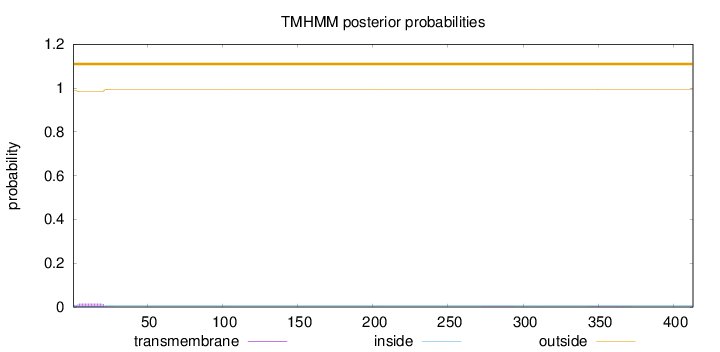
Population Genetic Test Statistics
Pi
25.023435
Theta
21.338338
Tajima's D
0.7756
CLR
0.819577
CSRT
0.601869906504675
Interpretation
Uncertain
