Pre Gene Modal
BGIBMGA010376
Annotation
uncharacterized_protein_LOC100141456_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.159
Sequence
CDS
ATGGATACCGTTGTGAAAAGCCTCCTAGAAGAAGATGAAGAAGAAAAAAAGGGTCTGGTCATATTTGGTAGAGACATATCAAAAATACCTTGCTTCAGAGAGAGCTTTCTTTATGGTATTGGTACTGGAATTGGATTTGGATTAGCCGCATTTGTGAAAACATCGAAGCCAATGCTAGCACAACACATAGGTGTTGGCACCTTCTCCCTGTCAACACTCTTATACTGGTCCTACTGCAGATACAGGTGGTCACAACAAAGGTTTGACGCACAGTTGTTACAAGATGCCTTAAAAGATAAAATTTTGTATGAAGGCACTGCAGTAGAAAAGGAAATGGAGAAAAAAGGTGTCTTAAAGACTGTATAG
Protein
MDTVVKSLLEEDEEEKKGLVIFGRDISKIPCFRESFLYGIGTGIGFGLAAFVKTSKPMLAQHIGVGTFSLSTLLYWSYCRYRWSQQRFDAQLLQDALKDKILYEGTAVEKEMEKKGVLKTV
Summary
Similarity
Belongs to the UDP-glycosyltransferase family.
Uniprot
O44232
H9JLH5
A0A2A4K9F0
A0A2H1V193
S4P827
A0A1E1VY22
+ More
A0A212FAA0 I4DLA1 A0A194PHJ6 A0A3S2TTG0 A0A0L7KQF7 A0A194R0E4 A0A2M3ZC38 A0A1Q3FFJ0 A0A2M4AX97 A0A2M4AX18 A0A182F4M5 W5J6T7 A0A2M4C3Z3 A0A2M4C3H2 A0A2M4C450 A0A1A9X1B9 T1DQZ4 A0A1L8DZX4 U5ESX7 A0A1L8EBQ5 A0A1L8EBL9 A0A1L8EDE5 A0A1L8ED65 A0A1B0CEL2 B0WBN7 A0A182LT21 A0A1I8P0J5 A0A0T6B968 Q16G81 A0A023EDY4 A0A182NE68 A0A0L0CJ83 A0A182K339 A7UUS4 A0A182HVB5 A0A182U7X5 A0A182WTC2 A0A182V939 A0A182T1K1 A0A182RBQ3 A0A182W9S9 A0A336MEV2 A0A1B0D5Q5 B4JEW7 A0A1A9XI28 A0A1B0G853 B4K8I4 B4M3U7 A0A3B0KVD6 A0A0P4VGS0 A0A1W4XVR6 B5DVG9 B4G5K7 A0A1J1HE03 A0A1W4UJA4 A0A1I8M726 A0A1B6FFS6 A0A0M4F6S8 B4N9Y5 B3P194 B4PPV5 A0A0A1WRQ4 Q9VG00 A0A0P4VTP3 B3LW48 A0A034WT72 A0A0K8UHI5 B4HGC2 B4R1X5 A0A1B6JHL1 A0A0V0G8N0 A0A067R2Z0 A0A224Y239 A0A069DP40 D6X1R2 A0A1A9VR22 K7IW71 A0A023F0F2 T1HCK4 A0A232FDL0 A0A1B6MAQ3 A0A1B6E2A4 N6U0B6
A0A212FAA0 I4DLA1 A0A194PHJ6 A0A3S2TTG0 A0A0L7KQF7 A0A194R0E4 A0A2M3ZC38 A0A1Q3FFJ0 A0A2M4AX97 A0A2M4AX18 A0A182F4M5 W5J6T7 A0A2M4C3Z3 A0A2M4C3H2 A0A2M4C450 A0A1A9X1B9 T1DQZ4 A0A1L8DZX4 U5ESX7 A0A1L8EBQ5 A0A1L8EBL9 A0A1L8EDE5 A0A1L8ED65 A0A1B0CEL2 B0WBN7 A0A182LT21 A0A1I8P0J5 A0A0T6B968 Q16G81 A0A023EDY4 A0A182NE68 A0A0L0CJ83 A0A182K339 A7UUS4 A0A182HVB5 A0A182U7X5 A0A182WTC2 A0A182V939 A0A182T1K1 A0A182RBQ3 A0A182W9S9 A0A336MEV2 A0A1B0D5Q5 B4JEW7 A0A1A9XI28 A0A1B0G853 B4K8I4 B4M3U7 A0A3B0KVD6 A0A0P4VGS0 A0A1W4XVR6 B5DVG9 B4G5K7 A0A1J1HE03 A0A1W4UJA4 A0A1I8M726 A0A1B6FFS6 A0A0M4F6S8 B4N9Y5 B3P194 B4PPV5 A0A0A1WRQ4 Q9VG00 A0A0P4VTP3 B3LW48 A0A034WT72 A0A0K8UHI5 B4HGC2 B4R1X5 A0A1B6JHL1 A0A0V0G8N0 A0A067R2Z0 A0A224Y239 A0A069DP40 D6X1R2 A0A1A9VR22 K7IW71 A0A023F0F2 T1HCK4 A0A232FDL0 A0A1B6MAQ3 A0A1B6E2A4 N6U0B6
Pubmed
9927176
19121390
23622113
22118469
22651552
26354079
+ More
26227816 20920257 23761445 17510324 24945155 26108605 12364791 14747013 17210077 17994087 27129103 15632085 25315136 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 24845553 26334808 18362917 19820115 20075255 25474469 28648823 23537049
26227816 20920257 23761445 17510324 24945155 26108605 12364791 14747013 17210077 17994087 27129103 15632085 25315136 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 24845553 26334808 18362917 19820115 20075255 25474469 28648823 23537049
EMBL
AB010579
BAA24526.1
BABH01005770
NWSH01000030
PCG80528.1
ODYU01000219
+ More
SOQ34620.1 GAIX01006131 JAA86429.1 GDQN01011429 JAT79625.1 AGBW02009504 OWR50666.1 AK402069 BAM18691.1 KQ459603 KPI92787.1 RSAL01000003 RVE54766.1 JTDY01007319 KOB65301.1 KQ460883 KPJ11172.1 GGFM01005385 MBW26136.1 GFDL01008729 JAV26316.1 GGFK01012011 MBW45332.1 GGFK01011990 MBW45311.1 ADMH02001962 ETN60172.1 GGFJ01010730 MBW59871.1 GGFJ01010729 MBW59870.1 GGFJ01010904 MBW60045.1 GAMD01000875 JAB00716.1 GFDF01002138 JAV11946.1 GANO01003024 JAB56847.1 GFDG01002619 JAV16180.1 GFDG01002784 JAV16015.1 GFDG01002117 JAV16682.1 GFDG01002119 JAV16680.1 AJWK01008981 DS231880 EDS42572.1 AXCM01000141 LJIG01009033 KRT83889.1 CH478315 EAT33244.1 GAPW01006081 GEHC01000233 JAC07517.1 JAV47412.1 JRES01000321 KNC32311.1 AAAB01008964 EDO63609.1 APCN01002504 UFQS01000916 UFQS01001101 UFQT01000916 UFQT01001101 SSX07636.1 SSX28922.1 AJVK01025510 CH916369 EDV93248.1 CCAG010000199 CH933806 EDW14383.1 CH940652 EDW59308.1 OUUW01000013 SPP87938.1 GDKW01003080 JAI53515.1 CM000070 EDY67915.1 CH479179 EDW24873.1 CVRI01000001 CRK86175.1 GECZ01022051 GECZ01020710 JAS47718.1 JAS49059.1 CP012526 ALC47457.1 CH964232 EDW81740.2 CH954181 EDV49283.1 CM000160 EDW97182.1 GBXI01013092 JAD01200.1 AE014297 AY119283 KX531864 AAF54894.1 AAM51143.1 ANY27674.1 GDRN01097619 GDRN01097612 GDRN01097610 GDRN01097609 JAI59101.1 CH902617 EDV42626.1 GAKP01001410 JAC57542.1 GDHF01026162 JAI26152.1 CH480815 EDW42370.1 CM000364 EDX13127.1 GECU01009059 JAS98647.1 GECL01001736 JAP04388.1 KK852736 KDR17422.1 GFTR01001927 JAW14499.1 GBGD01003532 JAC85357.1 KQ971371 EFA10149.1 GBBI01004266 JAC14446.1 ACPB03003258 NNAY01000414 OXU28583.1 GEBQ01007043 JAT32934.1 GEDC01005271 JAS32027.1 APGK01044277 KB741022 ENN74980.1
SOQ34620.1 GAIX01006131 JAA86429.1 GDQN01011429 JAT79625.1 AGBW02009504 OWR50666.1 AK402069 BAM18691.1 KQ459603 KPI92787.1 RSAL01000003 RVE54766.1 JTDY01007319 KOB65301.1 KQ460883 KPJ11172.1 GGFM01005385 MBW26136.1 GFDL01008729 JAV26316.1 GGFK01012011 MBW45332.1 GGFK01011990 MBW45311.1 ADMH02001962 ETN60172.1 GGFJ01010730 MBW59871.1 GGFJ01010729 MBW59870.1 GGFJ01010904 MBW60045.1 GAMD01000875 JAB00716.1 GFDF01002138 JAV11946.1 GANO01003024 JAB56847.1 GFDG01002619 JAV16180.1 GFDG01002784 JAV16015.1 GFDG01002117 JAV16682.1 GFDG01002119 JAV16680.1 AJWK01008981 DS231880 EDS42572.1 AXCM01000141 LJIG01009033 KRT83889.1 CH478315 EAT33244.1 GAPW01006081 GEHC01000233 JAC07517.1 JAV47412.1 JRES01000321 KNC32311.1 AAAB01008964 EDO63609.1 APCN01002504 UFQS01000916 UFQS01001101 UFQT01000916 UFQT01001101 SSX07636.1 SSX28922.1 AJVK01025510 CH916369 EDV93248.1 CCAG010000199 CH933806 EDW14383.1 CH940652 EDW59308.1 OUUW01000013 SPP87938.1 GDKW01003080 JAI53515.1 CM000070 EDY67915.1 CH479179 EDW24873.1 CVRI01000001 CRK86175.1 GECZ01022051 GECZ01020710 JAS47718.1 JAS49059.1 CP012526 ALC47457.1 CH964232 EDW81740.2 CH954181 EDV49283.1 CM000160 EDW97182.1 GBXI01013092 JAD01200.1 AE014297 AY119283 KX531864 AAF54894.1 AAM51143.1 ANY27674.1 GDRN01097619 GDRN01097612 GDRN01097610 GDRN01097609 JAI59101.1 CH902617 EDV42626.1 GAKP01001410 JAC57542.1 GDHF01026162 JAI26152.1 CH480815 EDW42370.1 CM000364 EDX13127.1 GECU01009059 JAS98647.1 GECL01001736 JAP04388.1 KK852736 KDR17422.1 GFTR01001927 JAW14499.1 GBGD01003532 JAC85357.1 KQ971371 EFA10149.1 GBBI01004266 JAC14446.1 ACPB03003258 NNAY01000414 OXU28583.1 GEBQ01007043 JAT32934.1 GEDC01005271 JAS32027.1 APGK01044277 KB741022 ENN74980.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000037510
+ More
UP000053240 UP000069272 UP000000673 UP000091820 UP000092461 UP000002320 UP000075883 UP000095300 UP000008820 UP000075884 UP000037069 UP000075881 UP000007062 UP000075840 UP000075902 UP000076407 UP000075903 UP000075901 UP000075900 UP000075920 UP000092462 UP000001070 UP000092443 UP000092444 UP000009192 UP000008792 UP000268350 UP000192223 UP000001819 UP000008744 UP000183832 UP000192221 UP000095301 UP000092553 UP000007798 UP000008711 UP000002282 UP000000803 UP000007801 UP000001292 UP000000304 UP000027135 UP000007266 UP000078200 UP000002358 UP000015103 UP000215335 UP000019118
UP000053240 UP000069272 UP000000673 UP000091820 UP000092461 UP000002320 UP000075883 UP000095300 UP000008820 UP000075884 UP000037069 UP000075881 UP000007062 UP000075840 UP000075902 UP000076407 UP000075903 UP000075901 UP000075900 UP000075920 UP000092462 UP000001070 UP000092443 UP000092444 UP000009192 UP000008792 UP000268350 UP000192223 UP000001819 UP000008744 UP000183832 UP000192221 UP000095301 UP000092553 UP000007798 UP000008711 UP000002282 UP000000803 UP000007801 UP000001292 UP000000304 UP000027135 UP000007266 UP000078200 UP000002358 UP000015103 UP000215335 UP000019118
Pfam
Interpro
IPR022533
Cox20
+ More
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR014307 Xanthine_DH_ssu
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR017978 GPCR_3_C
IPR036318 FAD-bd_PCMH-like_sf
IPR004122 BAF_prot
IPR036617 BAF_sf
IPR012337 RNaseH-like_sf
IPR002156 RNaseH_domain
IPR036397 RNaseH_sf
IPR000477 RT_dom
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR014307 Xanthine_DH_ssu
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR017978 GPCR_3_C
IPR036318 FAD-bd_PCMH-like_sf
IPR004122 BAF_prot
IPR036617 BAF_sf
SUPFAM
Gene 3D
CDD
ProteinModelPortal
O44232
H9JLH5
A0A2A4K9F0
A0A2H1V193
S4P827
A0A1E1VY22
+ More
A0A212FAA0 I4DLA1 A0A194PHJ6 A0A3S2TTG0 A0A0L7KQF7 A0A194R0E4 A0A2M3ZC38 A0A1Q3FFJ0 A0A2M4AX97 A0A2M4AX18 A0A182F4M5 W5J6T7 A0A2M4C3Z3 A0A2M4C3H2 A0A2M4C450 A0A1A9X1B9 T1DQZ4 A0A1L8DZX4 U5ESX7 A0A1L8EBQ5 A0A1L8EBL9 A0A1L8EDE5 A0A1L8ED65 A0A1B0CEL2 B0WBN7 A0A182LT21 A0A1I8P0J5 A0A0T6B968 Q16G81 A0A023EDY4 A0A182NE68 A0A0L0CJ83 A0A182K339 A7UUS4 A0A182HVB5 A0A182U7X5 A0A182WTC2 A0A182V939 A0A182T1K1 A0A182RBQ3 A0A182W9S9 A0A336MEV2 A0A1B0D5Q5 B4JEW7 A0A1A9XI28 A0A1B0G853 B4K8I4 B4M3U7 A0A3B0KVD6 A0A0P4VGS0 A0A1W4XVR6 B5DVG9 B4G5K7 A0A1J1HE03 A0A1W4UJA4 A0A1I8M726 A0A1B6FFS6 A0A0M4F6S8 B4N9Y5 B3P194 B4PPV5 A0A0A1WRQ4 Q9VG00 A0A0P4VTP3 B3LW48 A0A034WT72 A0A0K8UHI5 B4HGC2 B4R1X5 A0A1B6JHL1 A0A0V0G8N0 A0A067R2Z0 A0A224Y239 A0A069DP40 D6X1R2 A0A1A9VR22 K7IW71 A0A023F0F2 T1HCK4 A0A232FDL0 A0A1B6MAQ3 A0A1B6E2A4 N6U0B6
A0A212FAA0 I4DLA1 A0A194PHJ6 A0A3S2TTG0 A0A0L7KQF7 A0A194R0E4 A0A2M3ZC38 A0A1Q3FFJ0 A0A2M4AX97 A0A2M4AX18 A0A182F4M5 W5J6T7 A0A2M4C3Z3 A0A2M4C3H2 A0A2M4C450 A0A1A9X1B9 T1DQZ4 A0A1L8DZX4 U5ESX7 A0A1L8EBQ5 A0A1L8EBL9 A0A1L8EDE5 A0A1L8ED65 A0A1B0CEL2 B0WBN7 A0A182LT21 A0A1I8P0J5 A0A0T6B968 Q16G81 A0A023EDY4 A0A182NE68 A0A0L0CJ83 A0A182K339 A7UUS4 A0A182HVB5 A0A182U7X5 A0A182WTC2 A0A182V939 A0A182T1K1 A0A182RBQ3 A0A182W9S9 A0A336MEV2 A0A1B0D5Q5 B4JEW7 A0A1A9XI28 A0A1B0G853 B4K8I4 B4M3U7 A0A3B0KVD6 A0A0P4VGS0 A0A1W4XVR6 B5DVG9 B4G5K7 A0A1J1HE03 A0A1W4UJA4 A0A1I8M726 A0A1B6FFS6 A0A0M4F6S8 B4N9Y5 B3P194 B4PPV5 A0A0A1WRQ4 Q9VG00 A0A0P4VTP3 B3LW48 A0A034WT72 A0A0K8UHI5 B4HGC2 B4R1X5 A0A1B6JHL1 A0A0V0G8N0 A0A067R2Z0 A0A224Y239 A0A069DP40 D6X1R2 A0A1A9VR22 K7IW71 A0A023F0F2 T1HCK4 A0A232FDL0 A0A1B6MAQ3 A0A1B6E2A4 N6U0B6
Ontologies
GO
PANTHER
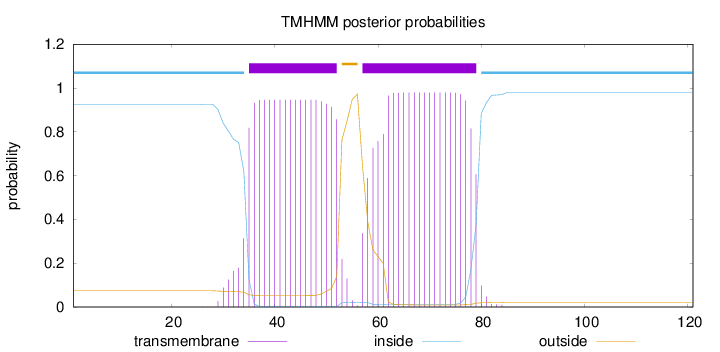
Topology
Length:
121
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.43658
Exp number, first 60 AAs:
20.43798
Total prob of N-in:
0.92629
POSSIBLE N-term signal
sequence
inside
1 - 34
TMhelix
35 - 52
outside
53 - 56
TMhelix
57 - 79
inside
80 - 121
Population Genetic Test Statistics
Pi
17.17469
Theta
18.582791
Tajima's D
-0.227169
CLR
0
CSRT
0.312084395780211
Interpretation
Uncertain
