Gene
KWMTBOMO07215 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010377
Annotation
xanthine_dehydrogenase_1_[Bombyx_mori]
Full name
Xanthine dehydrogenase
Alternative Name
Protein rosy locus
Location in the cell
Cytoplasmic Reliability : 1.447
Sequence
CDS
ATGGGATTATTGAATGCCGAGGAAGATCCGAATAAAATATGCACCGAACTTGTTTTCTACGTAAATGGTAAAAAGGTTATAGAATCTAGTCCTGATCCAGAATGGACATTATTATGGTATTTGCGAAAGAAACTTCGACTGACGGGCACAAAATTAGGTTGTGCGGAAGGTGGATGTGGTGCTTGCACTGTTATGGTCTCCAAATATAACCGACAAGAAAATAAAATTATTCATCTGGCAGTAAATGCTTGTCTCGCACCATTATGTGCTATGCACGGTTTGGCTGTGACCACAGTTGAAGGTATCGGATCAACCAAAACTAAGCTGCATCCAGTTCAAGAAAGAATAGCCAAAGCCCACGGATCACAGTGCGGTTTCTGTACGCCTGGAATCGTAATGTCCATGTATACTTTACTGAGAAGCTGTAAAAATATTCAATATTCTGATTTGGAGGTCGCGTTTCAAGGTAATTTATGTAGGTGTACAGGTTATCGCGCCATAATAGAGGGCTACAAAACGTTTATCGAAGATTGGGAGACTCAACGAATTGTCAAGAATGGACCACAGAACGGAACTTGTGCGATGGGGAAAGACTGTTGCAAAAATAAATCGGATAGCTGTGAAGAAGCAGATAGTGAATCACAATATATTTTTGACAAGTCGTCGTTCTTACCGTACGATTCCAGTCAAGAACCAATTTTTCCACCGGAATTGAAATTATCTTCAATATACGATAGTCAGTACGTAATATATCGTGGAAAACAAACAACGTGGTATCGGCCAACTAACATAGAAACAGTTTTATCGTTGAAAGATAAGTTCCCGAATGCAAAAGTGGTAGTGGGAAATAGTGAAGTTGGTGTTGAAGTTAAATTCAAACGTTGTGTTTATCCAATAATAATAATGCCTAATTGCGTTCCCGAATTGAATACTATTACTGAAAATGAGCATGGTTTAACGGTTGGAGCTTCAGTAACACTCAATGACATCGAAAAAACTTTTAGAGAGTATATAAAGAAACTACCACCATACAAAACTAGAGTCTTAACTGCTATAGTAGAAATGTTGAATTGGTTTGCTGGAAAACAAATCAGAAATGTTGCTGCCATTGGCGGGAATGTAATGACGGGCAGTCCAATATCTGATTTGAATCCGATTTTAATGTCTCTCAAGGTGAAACTTAATCTTCTCAGTCAAGAAAACGGCCATCGTACCGTGCTAATGGATGAAACTTTTTTCACTGGGTATAGAAAGAATGTCGTCAAATCAAATGAAATATTACTTTCAATAGAGATACCGTTTTCAACCAAATTTCAATACCTGAAGGCTATTAAGCAAGCGAAAAGAAGAGAGGACGATATATCCATAGTAACTTCAGCTGTTAATGTTGAATTCGAAGAGAACACAAATGTTATAAAATATATAAATTTGGCATTCGGGGGAATGGCTCCCGTAACAAAGATTGCAACAAACACTGGAAATGTGTTAAAAGGGCTTAAATGGAATGAAAACATGTTAGAAAAAGCGTATTCGTTACTGATAGATGAACTGCCATTAGATCCATCGGCACCAGGTGGAAATATACAATTTCGGCGTGCATTGACTATGAGTTTGTTTTTAAAGTCGTATTTGGCTATTGGAAAAGCAATGTCAACTGATTATTTTTACGGAGATCTTATTGAATCGTACTACGGTAGCGGTGCAGATAGTTTTCACGGAAATGTGCCGAAAAGTTCTCAATACTTTGAATTGGTCGGAGAAAAACAGCTTAAAAGTGATGCTGTTGGAAGGCCGATACAGCATATGTCAGCATACAAGCAAGCTACAGGCGAAGCAATATATTGCGATGATATGCCCATTGCAGAAGGTGAGCTATATTTAGCATTTGTTTTGAGTTCAAAAGCTCATGCAAAATTAATATCAGTAGATGCAAAAAAAGCGCTAGCCGAACCTGGTGTTATAGCATTTTATTCAGCAAAAGACTTAACAAAAGAACAAAATAGCATTGGCCCCATATTTCACGATGAAGAATTGTTTGCAAGAGATAAAGTGTTGAGTCAAGGTCAAACTATTGGCGTTATAGTAGCTGTGGATCAAGCGACTGCACAAGCTGCTGCAAGAATGGTTAAAGTTGAATACGAGGAGATACAGCCAATTATCGTTACTATAGAAGACGCTATAAAATATAACTCATTTTATCCACAATTCCCTAAAACAATAAAACGGGGAAATGTAAAAGCCGTGTTTGACGATAAAAATAATATTATAATAGAAGGACAGTGTCGTATGGGAGGTCAAGAACATTTTTATTTGGAGACTCATGCTGCGTTTGCTATTCCAAAGAAAGAAGACGACGAGCTAGAAATATTTTGTTCCAGCCAACATCCCTCAGAGATCGCAAAACTGGTTAGTCACATCCTGCACGTACCGATGAATAGAATAGTGGCGAGAGTGAAACGTATGGGTGGTGGTTTTGGTGGAAAAGAGTCACGGGGTATGTTAGTGGCATTACCGGTTGCATTAGCTGCTCATAAACTCAACAGGCCCGTGCGGTGCATGCTCGATAGAGACGAAGATATGCAGATGACGGGCACTAGACATCCATTCCTGATAAAGTATAAAGCAGCGGCAACTAAAGAAGGAAAAATTGTTGGAGCTGTTGTCAATATTTATAACAACGGAGGCTATTCCACTGATCTGTCAGGTCCAGTTGTCGAACGAGCAATGTTCCATTTTGAAAATGCTTATTACATCCCAAATTGTGAAGTGACCGGATATGTCTGTAGAACTAATTTACCCTCAAATACGGCTTTTCGAGGCTTCGGAGGTCCCCAAGGAATGTTTGGTGCCGAAAATATGGTTAGAGAGATAGCTCATCGACTGGGAAAATCTCCCGAGGAAATAAGCAGACTAAACTTGTATCGAGAAAATAATACTACACATTATGGACAAGTTCTGACATATTGTACGTTACAAAGATGTTGGGACGAATGTGTACAAAACTGTAATTTAGCAGAAAGAAAATTAAAAATTAAAGAATTCAACAAACAACATAGATGGAGAAAACGTGGTATTTCGATAATACCAACAAAATTCGGTATCGCTTTTACGGAGAAACTTTTAAACCAAGCTGGTGCATTGGTATTAGTTTATGTTGATGGTTCAGTACTTCTATCACACGGTGGTACTGAGATGGGACAGGGCTTACATACAAAGATGATACAAGTAGCTACTCGGGCTCTTGGTATAGACGTTTCTAAAATTCACATCAGCGAAACAAGTACAGATAAAGTCCCTAATACTTCGGCAACAGCGGCTAGTGCCGGCTCAGATTTGAACGGTATGGCAGTACTAGAGGCTTGCGAGAAAATAATGAAGCGCCTAAAACCGTATATAGATAAGAATCCGGACGGGAAATGGGAAAATTGGGTATCAGCTGCCTATGTCGACCGTGTTAGCTTATCCGCTACCGGCTTCCACGCTACACCGGATATAGGATTCGATTTCAAGACCACTTCGGGAAAACCGTTTAACTATTTCACATACGGTGTTGCTTGCACGGAAGTAGAAATTGATTGTCTTAGCGGTGACCACCAAGTCCTTAGAACGGATATTGTAATGGACCTCGGTGAGAGTATCAATCCAGCGATAGATATTGGCCAGATTGAAGGCGGTTTCATACAAGGCTACGGTTTGTTTACCATAGAAGAATTAATTTATTCACCCACCGGAACTTTATATTCGCGAGGGCCGGGTGCTTACAAAATTCCTGGATTTGGCGATATTCCTCTAGAATTCAACGTTTCATTGTTAAAGGGTGCACCTAACCCTCGAGCAGTATATTCTTCTAAGGCCGTAGGAGAACCTCCGCTGTTCTTGGCTTCTTCAGCTTATTTTGCGATACATGAAGCCATTAAAGCTGCAAGAGCAGATGCCGGAGTACCGCTCGAGTTCGACATGGAGGCACCAGCAACATCTGCTCGAATTCGAATGGCATGTGAGGACCATATAACGAAGAAACTTGATAAACCAGATCCAAATAGCTTTGTGCCGTGGAATGTTGTCCCTTAA
Protein
MGLLNAEEDPNKICTELVFYVNGKKVIESSPDPEWTLLWYLRKKLRLTGTKLGCAEGGCGACTVMVSKYNRQENKIIHLAVNACLAPLCAMHGLAVTTVEGIGSTKTKLHPVQERIAKAHGSQCGFCTPGIVMSMYTLLRSCKNIQYSDLEVAFQGNLCRCTGYRAIIEGYKTFIEDWETQRIVKNGPQNGTCAMGKDCCKNKSDSCEEADSESQYIFDKSSFLPYDSSQEPIFPPELKLSSIYDSQYVIYRGKQTTWYRPTNIETVLSLKDKFPNAKVVVGNSEVGVEVKFKRCVYPIIIMPNCVPELNTITENEHGLTVGASVTLNDIEKTFREYIKKLPPYKTRVLTAIVEMLNWFAGKQIRNVAAIGGNVMTGSPISDLNPILMSLKVKLNLLSQENGHRTVLMDETFFTGYRKNVVKSNEILLSIEIPFSTKFQYLKAIKQAKRREDDISIVTSAVNVEFEENTNVIKYINLAFGGMAPVTKIATNTGNVLKGLKWNENMLEKAYSLLIDELPLDPSAPGGNIQFRRALTMSLFLKSYLAIGKAMSTDYFYGDLIESYYGSGADSFHGNVPKSSQYFELVGEKQLKSDAVGRPIQHMSAYKQATGEAIYCDDMPIAEGELYLAFVLSSKAHAKLISVDAKKALAEPGVIAFYSAKDLTKEQNSIGPIFHDEELFARDKVLSQGQTIGVIVAVDQATAQAAARMVKVEYEEIQPIIVTIEDAIKYNSFYPQFPKTIKRGNVKAVFDDKNNIIIEGQCRMGGQEHFYLETHAAFAIPKKEDDELEIFCSSQHPSEIAKLVSHILHVPMNRIVARVKRMGGGFGGKESRGMLVALPVALAAHKLNRPVRCMLDRDEDMQMTGTRHPFLIKYKAAATKEGKIVGAVVNIYNNGGYSTDLSGPVVERAMFHFENAYYIPNCEVTGYVCRTNLPSNTAFRGFGGPQGMFGAENMVREIAHRLGKSPEEISRLNLYRENNTTHYGQVLTYCTLQRCWDECVQNCNLAERKLKIKEFNKQHRWRKRGISIIPTKFGIAFTEKLLNQAGALVLVYVDGSVLLSHGGTEMGQGLHTKMIQVATRALGIDVSKIHISETSTDKVPNTSATAASAGSDLNGMAVLEACEKIMKRLKPYIDKNPDGKWENWVSAAYVDRVSLSATGFHATPDIGFDFKTTSGKPFNYFTYGVACTEVEIDCLSGDHQVLRTDIVMDLGESINPAIDIGQIEGGFIQGYGLFTIEELIYSPTGTLYSRGPGAYKIPGFGDIPLEFNVSLLKGAPNPRAVYSSKAVGEPPLFLASSAYFAIHEAIKAARADAGVPLEFDMEAPATSARIRMACEDHITKKLDKPDPNSFVPWNVVP
Summary
Description
Key enzyme in purine degradation. Catalyzes the oxidation of hypoxanthine to xanthine. Catalyzes the oxidation of xanthine to uric acid (By similarity).
Catalytic Activity
H2O + NAD(+) + xanthine = H(+) + NADH + urate
H2O + hypoxanthine + NAD(+) = H(+) + NADH + xanthine
H2O + hypoxanthine + NAD(+) = H(+) + NADH + xanthine
Cofactor
FAD
Mo-molybdopterin
[2Fe-2S] cluster
Mo-molybdopterin
[2Fe-2S] cluster
Subunit
Homodimer.
Similarity
Belongs to the xanthine dehydrogenase family.
Belongs to the UDP-glycosyltransferase family.
Belongs to the UDP-glycosyltransferase family.
Keywords
2Fe-2S
Complete proteome
FAD
Flavoprotein
Iron
Iron-sulfur
Metal-binding
Molybdenum
NAD
Oxidoreductase
Peroxisome
Reference proteome
Feature
chain Xanthine dehydrogenase
Uniprot
O17506
Q17209
H9JLH6
A0A2H1V2N7
A0A3S2NRU2
A0A2A4K8J0
+ More
A0A212FAB6 A0A194R1Q5 A0A0L7KUC5 A0A194PNT2 A0A1Y1M1D6 A0A0V0G4W5 A0A0A9WB52 A0A151IP28 A0A069DYC1 A0A158NVN6 A0A195EC39 A0A1B6CHR7 A0A224XCZ2 A0A3L8D698 A0A067RHQ5 A0A195FXD8 A0A195B8M6 A0A151XC24 A0A232FH45 F4WNQ7 A8E1U4 A0A182GI29 A0A2M4A109 A0A0N0BEE7 A0A2M4A100 A0A1S4F2J6 Q17HF7 A0A0C9R3C5 A0A1B6EAL5 T1I5G4 A0A2M4A1L3 A0A087ZV59 A0A2M3Z169 A0A0B4J2Q7 A0A2M4BA61 A0A1L8DYE4 W5JBD8 A0A182F4M3 A0A182NE47 A0A2M4B9K9 E2BWN6 A0A2R7WTC8 A0A154P411 A0A1S4JBA4 A0A0B4J2Q6 A0A1B6C366 A0A182UMJ7 A0A1Q3FH76 A0A336MDC5 A0A2A3E831 A0A336MH61 A0A1S4GWN9 B0WBP0 A0A1W4UJ58 P10351 B4PPV6 B3P193 P08793 A0A1L2FW41 B4HGC1 B3LW47 A0A0L7R1H8 B4K8I3 B4G5K8 A0A182J3B6 A0A0L0CIT4 P22811 B4N9Y6 Q6WMV5 Q6WMV4 Q6WMV1 W4VRL6 B4JEW6 A0A3B0K0X8 P91711 Q6WMV6 Q23829 A0A0M4EK34 Q6WMV3 Q7Q3J8 E0VM07 Q6WMV0 B4M3U6 Q6WMV2 Q9BIF9 Q6WMU9 A0A1L2FW42 A0A0K8U3V6 W8BAW7 T1PDW6 A0A1I8PGD3 A0A1J1HE03 A0A1I8MT78 A0A2H1V1C3
A0A212FAB6 A0A194R1Q5 A0A0L7KUC5 A0A194PNT2 A0A1Y1M1D6 A0A0V0G4W5 A0A0A9WB52 A0A151IP28 A0A069DYC1 A0A158NVN6 A0A195EC39 A0A1B6CHR7 A0A224XCZ2 A0A3L8D698 A0A067RHQ5 A0A195FXD8 A0A195B8M6 A0A151XC24 A0A232FH45 F4WNQ7 A8E1U4 A0A182GI29 A0A2M4A109 A0A0N0BEE7 A0A2M4A100 A0A1S4F2J6 Q17HF7 A0A0C9R3C5 A0A1B6EAL5 T1I5G4 A0A2M4A1L3 A0A087ZV59 A0A2M3Z169 A0A0B4J2Q7 A0A2M4BA61 A0A1L8DYE4 W5JBD8 A0A182F4M3 A0A182NE47 A0A2M4B9K9 E2BWN6 A0A2R7WTC8 A0A154P411 A0A1S4JBA4 A0A0B4J2Q6 A0A1B6C366 A0A182UMJ7 A0A1Q3FH76 A0A336MDC5 A0A2A3E831 A0A336MH61 A0A1S4GWN9 B0WBP0 A0A1W4UJ58 P10351 B4PPV6 B3P193 P08793 A0A1L2FW41 B4HGC1 B3LW47 A0A0L7R1H8 B4K8I3 B4G5K8 A0A182J3B6 A0A0L0CIT4 P22811 B4N9Y6 Q6WMV5 Q6WMV4 Q6WMV1 W4VRL6 B4JEW6 A0A3B0K0X8 P91711 Q6WMV6 Q23829 A0A0M4EK34 Q6WMV3 Q7Q3J8 E0VM07 Q6WMV0 B4M3U6 Q6WMV2 Q9BIF9 Q6WMU9 A0A1L2FW42 A0A0K8U3V6 W8BAW7 T1PDW6 A0A1I8PGD3 A0A1J1HE03 A0A1I8MT78 A0A2H1V1C3
EC Number
1.17.1.4
Pubmed
9927176
9674379
19121390
22118469
26354079
26227816
+ More
28004739 25401762 26334808 21347285 30249741 24845553 28648823 21719571 18510977 26483478 17510324 20075255 20920257 23761445 20798317 12364791 10731132 12537572 3036645 3036646 12537569 17994087 17550304 2516831 2830167 26108605 2493563 15632085 15545653 8913749 20566863 11514452 24495485 25315136
28004739 25401762 26334808 21347285 30249741 24845553 28648823 21719571 18510977 26483478 17510324 20075255 20920257 23761445 20798317 12364791 10731132 12537572 3036645 3036646 12537569 17994087 17550304 2516831 2830167 26108605 2493563 15632085 15545653 8913749 20566863 11514452 24495485 25315136
EMBL
AB005911
BAA21640.1
D38159
BAA07348.1
BABH01005768
BABH01005769
+ More
BABH01005770 ODYU01000219 SOQ34622.1 RSAL01000003 RVE54763.1 NWSH01000030 PCG80531.1 AGBW02009504 OWR50668.1 KQ460883 KPJ11175.1 JTDY01005560 KOB66882.1 KQ459603 KPI92785.1 GEZM01042838 JAV79553.1 GECL01003376 JAP02748.1 GBHO01039856 GBRD01002792 JAG03748.1 JAG63029.1 KQ976892 KYN07230.1 GBGD01000107 JAC88782.1 ADTU01002895 KQ979074 KYN22790.1 GEDC01024291 JAS13007.1 GFTR01008778 JAW07648.1 QOIP01000012 RLU15975.1 KK852674 KDR18732.1 KQ981215 KYN44509.1 KQ976565 KYM80544.1 KQ982316 KYQ57848.1 NNAY01000242 OXU29769.1 GL888237 EGI64250.1 AM887864 CAP08999.1 JXUM01065174 JXUM01065175 KQ562333 KXJ76128.1 GGFK01001120 MBW34441.1 KQ435830 KOX71781.1 GGFK01001110 MBW34431.1 CH477249 EAT46105.1 GBYB01010764 JAG80531.1 GEDC01002348 JAS34950.1 ACPB03003258 GGFK01001318 MBW34639.1 GGFM01001519 MBW22270.1 GGFJ01000577 MBW49718.1 GFDF01002809 JAV11275.1 ADMH02001962 ETN60170.1 GGFJ01000576 MBW49717.1 GL451165 EFN79891.1 KK855492 PTY22814.1 KQ434804 KZC06074.1 GEDC01029337 JAS07961.1 GFDL01008163 JAV26882.1 UFQS01000746 UFQT01000746 SSX06562.1 SSX26909.1 KZ288336 PBC27858.1 UFQT01001227 SSX29566.1 AAAB01008964 DS231880 EDS42575.1 Y00308 AE014297 BT015293 AY094689 CM000160 EDW97183.1 CH954181 EDV49282.1 X07229 X07323 X07324 X07325 M18423 KX384737 AOE46690.1 CH480815 EDW42369.1 CH902617 EDV42625.1 KQ414668 KOC64646.1 CH933806 EDW14382.1 CH479179 EDW24874.1 JRES01000321 KNC32318.1 M33977 CM000070 AY754605 CH964232 EDW81741.1 AY279337 AAQ17527.1 AY279338 AAQ17528.1 AY279341 AAQ17531.1 GANO01001536 JAB58335.1 CH916369 EDV93247.1 OUUW01000013 SPP87937.1 Y08237 AY279336 AAQ17526.1 M30316 AAA27880.1 CP012526 ALC47458.1 AY279339 AAQ17529.1 EAA12866.3 DS235286 EEB14413.1 AY279342 AAQ17532.1 CH940652 EDW59307.1 AY279340 AAQ17530.1 AY014961 GAMC01016290 AAG47345.1 JAB90265.1 AY279343 AAQ17533.1 KX384734 AOE46687.1 GDHF01031091 JAI21223.1 GAMC01016289 JAB90266.1 KA646889 AFP61518.1 CVRI01000001 CRK86175.1 SOQ34621.1
BABH01005770 ODYU01000219 SOQ34622.1 RSAL01000003 RVE54763.1 NWSH01000030 PCG80531.1 AGBW02009504 OWR50668.1 KQ460883 KPJ11175.1 JTDY01005560 KOB66882.1 KQ459603 KPI92785.1 GEZM01042838 JAV79553.1 GECL01003376 JAP02748.1 GBHO01039856 GBRD01002792 JAG03748.1 JAG63029.1 KQ976892 KYN07230.1 GBGD01000107 JAC88782.1 ADTU01002895 KQ979074 KYN22790.1 GEDC01024291 JAS13007.1 GFTR01008778 JAW07648.1 QOIP01000012 RLU15975.1 KK852674 KDR18732.1 KQ981215 KYN44509.1 KQ976565 KYM80544.1 KQ982316 KYQ57848.1 NNAY01000242 OXU29769.1 GL888237 EGI64250.1 AM887864 CAP08999.1 JXUM01065174 JXUM01065175 KQ562333 KXJ76128.1 GGFK01001120 MBW34441.1 KQ435830 KOX71781.1 GGFK01001110 MBW34431.1 CH477249 EAT46105.1 GBYB01010764 JAG80531.1 GEDC01002348 JAS34950.1 ACPB03003258 GGFK01001318 MBW34639.1 GGFM01001519 MBW22270.1 GGFJ01000577 MBW49718.1 GFDF01002809 JAV11275.1 ADMH02001962 ETN60170.1 GGFJ01000576 MBW49717.1 GL451165 EFN79891.1 KK855492 PTY22814.1 KQ434804 KZC06074.1 GEDC01029337 JAS07961.1 GFDL01008163 JAV26882.1 UFQS01000746 UFQT01000746 SSX06562.1 SSX26909.1 KZ288336 PBC27858.1 UFQT01001227 SSX29566.1 AAAB01008964 DS231880 EDS42575.1 Y00308 AE014297 BT015293 AY094689 CM000160 EDW97183.1 CH954181 EDV49282.1 X07229 X07323 X07324 X07325 M18423 KX384737 AOE46690.1 CH480815 EDW42369.1 CH902617 EDV42625.1 KQ414668 KOC64646.1 CH933806 EDW14382.1 CH479179 EDW24874.1 JRES01000321 KNC32318.1 M33977 CM000070 AY754605 CH964232 EDW81741.1 AY279337 AAQ17527.1 AY279338 AAQ17528.1 AY279341 AAQ17531.1 GANO01001536 JAB58335.1 CH916369 EDV93247.1 OUUW01000013 SPP87937.1 Y08237 AY279336 AAQ17526.1 M30316 AAA27880.1 CP012526 ALC47458.1 AY279339 AAQ17529.1 EAA12866.3 DS235286 EEB14413.1 AY279342 AAQ17532.1 CH940652 EDW59307.1 AY279340 AAQ17530.1 AY014961 GAMC01016290 AAG47345.1 JAB90265.1 AY279343 AAQ17533.1 KX384734 AOE46687.1 GDHF01031091 JAI21223.1 GAMC01016289 JAB90266.1 KA646889 AFP61518.1 CVRI01000001 CRK86175.1 SOQ34621.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000037510
+ More
UP000053268 UP000078542 UP000005205 UP000078492 UP000279307 UP000027135 UP000078541 UP000078540 UP000075809 UP000215335 UP000007755 UP000069940 UP000249989 UP000053105 UP000008820 UP000015103 UP000005203 UP000002358 UP000000673 UP000069272 UP000075884 UP000008237 UP000076502 UP000075903 UP000242457 UP000002320 UP000192221 UP000000803 UP000002282 UP000008711 UP000001292 UP000007801 UP000053825 UP000009192 UP000008744 UP000075880 UP000037069 UP000001819 UP000007798 UP000001070 UP000268350 UP000092553 UP000007062 UP000009046 UP000008792 UP000095300 UP000183832 UP000095301
UP000053268 UP000078542 UP000005205 UP000078492 UP000279307 UP000027135 UP000078541 UP000078540 UP000075809 UP000215335 UP000007755 UP000069940 UP000249989 UP000053105 UP000008820 UP000015103 UP000005203 UP000002358 UP000000673 UP000069272 UP000075884 UP000008237 UP000076502 UP000075903 UP000242457 UP000002320 UP000192221 UP000000803 UP000002282 UP000008711 UP000001292 UP000007801 UP000053825 UP000009192 UP000008744 UP000075880 UP000037069 UP000001819 UP000007798 UP000001070 UP000268350 UP000092553 UP000007062 UP000009046 UP000008792 UP000095300 UP000183832 UP000095301
Pfam
Interpro
IPR001041
2Fe-2S_ferredoxin-type
+ More
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR002888 2Fe-2S-bd
IPR022407 OxRdtase_Mopterin_BS
IPR014307 Xanthine_DH_ssu
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR016167 FAD-bd_PCMH_sub1
IPR016169 FAD-bd_PCMH_sub2
IPR014309 Xanthine_DH_Mopterin-bd_su
IPR012675 Beta-grasp_dom_sf
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR001497 MethylDNA_cys_MeTrfase_AS
IPR022533 Cox20
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR017978 GPCR_3_C
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR002888 2Fe-2S-bd
IPR022407 OxRdtase_Mopterin_BS
IPR014307 Xanthine_DH_ssu
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR016167 FAD-bd_PCMH_sub1
IPR016169 FAD-bd_PCMH_sub2
IPR014309 Xanthine_DH_Mopterin-bd_su
IPR012675 Beta-grasp_dom_sf
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR001497 MethylDNA_cys_MeTrfase_AS
IPR022533 Cox20
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR017978 GPCR_3_C
SUPFAM
Gene 3D
CDD
ProteinModelPortal
O17506
Q17209
H9JLH6
A0A2H1V2N7
A0A3S2NRU2
A0A2A4K8J0
+ More
A0A212FAB6 A0A194R1Q5 A0A0L7KUC5 A0A194PNT2 A0A1Y1M1D6 A0A0V0G4W5 A0A0A9WB52 A0A151IP28 A0A069DYC1 A0A158NVN6 A0A195EC39 A0A1B6CHR7 A0A224XCZ2 A0A3L8D698 A0A067RHQ5 A0A195FXD8 A0A195B8M6 A0A151XC24 A0A232FH45 F4WNQ7 A8E1U4 A0A182GI29 A0A2M4A109 A0A0N0BEE7 A0A2M4A100 A0A1S4F2J6 Q17HF7 A0A0C9R3C5 A0A1B6EAL5 T1I5G4 A0A2M4A1L3 A0A087ZV59 A0A2M3Z169 A0A0B4J2Q7 A0A2M4BA61 A0A1L8DYE4 W5JBD8 A0A182F4M3 A0A182NE47 A0A2M4B9K9 E2BWN6 A0A2R7WTC8 A0A154P411 A0A1S4JBA4 A0A0B4J2Q6 A0A1B6C366 A0A182UMJ7 A0A1Q3FH76 A0A336MDC5 A0A2A3E831 A0A336MH61 A0A1S4GWN9 B0WBP0 A0A1W4UJ58 P10351 B4PPV6 B3P193 P08793 A0A1L2FW41 B4HGC1 B3LW47 A0A0L7R1H8 B4K8I3 B4G5K8 A0A182J3B6 A0A0L0CIT4 P22811 B4N9Y6 Q6WMV5 Q6WMV4 Q6WMV1 W4VRL6 B4JEW6 A0A3B0K0X8 P91711 Q6WMV6 Q23829 A0A0M4EK34 Q6WMV3 Q7Q3J8 E0VM07 Q6WMV0 B4M3U6 Q6WMV2 Q9BIF9 Q6WMU9 A0A1L2FW42 A0A0K8U3V6 W8BAW7 T1PDW6 A0A1I8PGD3 A0A1J1HE03 A0A1I8MT78 A0A2H1V1C3
A0A212FAB6 A0A194R1Q5 A0A0L7KUC5 A0A194PNT2 A0A1Y1M1D6 A0A0V0G4W5 A0A0A9WB52 A0A151IP28 A0A069DYC1 A0A158NVN6 A0A195EC39 A0A1B6CHR7 A0A224XCZ2 A0A3L8D698 A0A067RHQ5 A0A195FXD8 A0A195B8M6 A0A151XC24 A0A232FH45 F4WNQ7 A8E1U4 A0A182GI29 A0A2M4A109 A0A0N0BEE7 A0A2M4A100 A0A1S4F2J6 Q17HF7 A0A0C9R3C5 A0A1B6EAL5 T1I5G4 A0A2M4A1L3 A0A087ZV59 A0A2M3Z169 A0A0B4J2Q7 A0A2M4BA61 A0A1L8DYE4 W5JBD8 A0A182F4M3 A0A182NE47 A0A2M4B9K9 E2BWN6 A0A2R7WTC8 A0A154P411 A0A1S4JBA4 A0A0B4J2Q6 A0A1B6C366 A0A182UMJ7 A0A1Q3FH76 A0A336MDC5 A0A2A3E831 A0A336MH61 A0A1S4GWN9 B0WBP0 A0A1W4UJ58 P10351 B4PPV6 B3P193 P08793 A0A1L2FW41 B4HGC1 B3LW47 A0A0L7R1H8 B4K8I3 B4G5K8 A0A182J3B6 A0A0L0CIT4 P22811 B4N9Y6 Q6WMV5 Q6WMV4 Q6WMV1 W4VRL6 B4JEW6 A0A3B0K0X8 P91711 Q6WMV6 Q23829 A0A0M4EK34 Q6WMV3 Q7Q3J8 E0VM07 Q6WMV0 B4M3U6 Q6WMV2 Q9BIF9 Q6WMU9 A0A1L2FW42 A0A0K8U3V6 W8BAW7 T1PDW6 A0A1I8PGD3 A0A1J1HE03 A0A1I8MT78 A0A2H1V1C3
PDB
4YTY
E-value=0,
Score=3550
Ontologies
PATHWAY
GO
GO:0004854
GO:0043546
GO:0051537
GO:0005506
GO:0071949
GO:0009055
GO:0004855
GO:0016491
GO:0046872
GO:0030151
GO:0042626
GO:0005524
GO:0016021
GO:0006144
GO:0006525
GO:0006650
GO:0009115
GO:0006568
GO:0048072
GO:0050660
GO:0008340
GO:0006206
GO:0005777
GO:0005829
GO:0006281
GO:0003908
GO:0033617
GO:0009060
GO:0004930
GO:0016758
GO:0005743
GO:0051536
GO:0003824
GO:0016614
GO:0055114
Topology
Subcellular location
Peroxisome
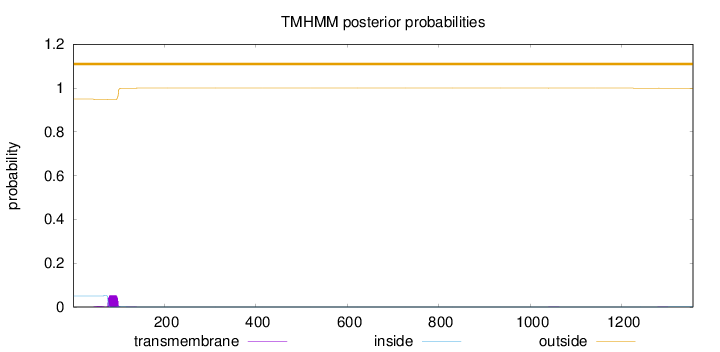
Length:
1356
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.26157000000001
Exp number, first 60 AAs:
0.03534
Total prob of N-in:
0.05018
outside
1 - 1356
Population Genetic Test Statistics
Pi
26.564183
Theta
21.109782
Tajima's D
0.252965
CLR
0.564871
CSRT
0.450927453627319
Interpretation
Uncertain
