Gene
KWMTBOMO07214 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010574
Annotation
Bm8_interacting_protein_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.658
Sequence
CDS
ATGACTGTAATAAAATTTATTTTAGTTTTAATATTTATAGCTCCATCATATGGTAGAGTGCCTGATAAAGTTTATTGTTTAGATGAAACATGCAGCAAGCCACTTTCAAGAGCCAAAACTTTGATAACTTACTCAAGTGCGGATCCTGACCACATATCCTATCGTAATAATGCTGAAGCTCTGGTGTTTATGAAGTCAGCTGGAACAAACCCTGAGCTATGGTTTGCAAGTATTGAAGTGCCAACAGAGTCATTGATACCAAAAATTTCCTTTCAACCTGATAAAGTTAAACAACCCCATGAAGTGGTCGAGGGTACCACACTGTACACTACGGAAGCAACAGTCAAAGATATCAATACAGCACCACAACCTGCATCAAGTTTACCAAATGAAATTAATGAAAATTTAGGTACAATTAAACCAGTTGACAATATTAACCCAAAAAATGTTGATTTTGAAAACATTAGAGAAGTCAACAAAAGGCTCTTAGATACTCCTAATTTTCCTCAACAAAAAATACCAAATGTTCAAGAAGCACAAGAGAATATTCAGAAAAAAAATACAATTGAAACAAATGTTCGATTTGAAAATGTACCAAATCAACATTTAAATGTTCATAAGGATGTCAATGTTCATAATCCAGAAACAAAAGATCAGAACTACTATAAACAAGTAAATTTGAATGAAGCAGAATTACAGAAAACAAGTGAAACATTGACAACAGTTCCAATACAAAAATTCAATGAAGAGAATTTTCATGATCACAACATTTTGAATGAGAAGTCAGTTCAAAAAGGGCATGAGCAGCATGTTAATGATTTACCTATTATAGAAAATAGTAGTCCTAACGTTCACAATAAAGAAGTTTATAATGAGCCTCAAATTTCCATCTCTCAATCTTCATCAAGAAGTGGCAGTGAAGGAAATATCCCTAATACAGAAGGCATTATTCATCAAATGCTAAACGATGAAGTCCCAGAAAGTGAACAAAATGTTTCAGTGCCTTCAAACATGAATAGAAATGAAGATAGTGATAAACATGCAAAACAAGCAGTAGCAGATAGTACTGTAGAAAATACACAACAACAGAACAATGTGTATAATAATAATCAAAAAGGGCATGAGCAGCATGTTAATGATTTACCTATTATAGAAAATAGCAGTCCTAACATTCACAATAAAGAAGTTTATAATGAGCCTCAAATTTCCATCTCTCAATCTTCGTCAAGAAGTGGCAGTGAAGGAAATATCCCTAATACGGAAGGCACTATTCATCAAATGCTAAACGATGAAGTCCCAGAAAGTGAACAAAATGTTTCAGTGCCTTCAAACATGAATAGAAATGAAGATCGTGATAAACATGCAAAACAAGCAGTAGCAGATAGTACTGTAGAAAATACACAACAACAGAACAATCTATATAATAATATTCCAAATATAAATGGGGAACAAGTTCAAAAAAACCCAGAGCTCCCATTAGAGACAAACAGAGAACAAAATAATTTCTTAGACCAACAAAATCACCCAAACGAAAAATTAAATAAAAATCATTTTGAAAAAAGTACACCTTTTATGGAAAACCCTTCTAATATTTTATATGACAATACTCAAAGTGAAATAAATAGGCTGACAGAAACAATTAGTACTAAGACAGAGATAGAATCCGAACGAACTTTGCCCATTGTAAATAGATCACCCAATTCTGATAATGTTGTAAACTATGAAACAGGAATACCATCTACTCTTAGTGTAACAGAGAGAAATATCGAAATAAATAACCCCAATACAGGAATGCCTCCAATTGTTGAAAATCCTAACAACATAATTCCTAACATAAATTTGAAGGAAATACAATTGGAATCAAGGGAGGTACCCTCAGTAGATAGATCTCACAGTCCTGAATATGTCTTAAACAATCAAAACGAAAATATACGAACACCACCCCATCCAGCTGAAACACAAAGATATTTTGAAACAAATAATGCCATGACTGAAATACCTCCAGTTGTTGAAAATGTTAACAATATTGTTCATAATGCGAACTTAAAGGAACAACAACTGGAATCAATACCTCCCTATGTAGACAGAGCTCCCAATCAAGAAAATGTATTTGTTGATGTGGCAGAACAATTAAGAACACCACCTCCTCCATATTTGAATCCCATAACTGAGAATTATCAACAAGATCATTTTGAAAATCCCACTACAGAAACGTCACCACTTCTATTTAATCCATCCACATCTGCAATACCCGATATTTCCATAAATGAAGAGCAACCATTAACAACAGAATTAAATATACAAACAACAACTGAAGGTTCAAAAGAACCATTTTCAGGCATGGATGAGAACACAAACATTAATCAGGAATTGCAAAGTAACCAGGAAGTTGGTGATAAACACTCCTACTACACGGAAACTCCTGTAGATATTGAATATGGTAACACAGCAGAGTCAAATTTACCAACTAGTGATGAATACCAGGTATTTCCTGAAGAAACCACTAAAGGATGGTTTGCTTCCATTTTTGACACATTAGCTGGTATGTGGACCTCATCATCTGATGATCAAGACATTTCAGATAATACTGAAGATAGCATCTATGATGAACCAAAGACTGACAATGCTGAAGAATTTTCTTTTATAAAATACTTAATACATAGTTATTATTCAGTGATGGGAATAGATGAAGAAACTCGAGTACTCTTCACATCTGCAGGAGACGCCTGCTATACAGAAAACTACTGTGATGGGCAAAGTAACACAAATAAAAATCGATTGTTAACATTCCTCCTAACGACAGCCAGCTCTGTTCTTCTGTTCACATTGGGATATTATTACATCGACATAAGGAGACAGGATGGTAGGCTTATTGGAACAATCAATTCTTTGCAAAGAGATTTGTTATTTACTACTAAGGAGTGTGAGATTTTAAAAGAGGAGCTTTCGACTACTAAGACGAAATTAGCAGGGATTGAAGACAACTCATTTGGCATGGACGATATGGTGCAATCACTAAAAGAGGAAATCAATGAAATGAAAGCACAGAACGATAGGCTACGTCATTCTCTGGACGACAATGAAAAACTGTTGCGAGTATCCGAAAATACTGCCGAGGAGCTTCAGAACACTCTCAGCGAAGTGGAGAATACTCTCAGTGAGCTTTTGGCTGAAAGAGCTAACTTAGAGGAGCAAATTGCAGAAATCAATGGAAAAGCTCAAGCATTTGAAGAGGAATTGATCACAGTAAGTCGAGATCGAGATAACTTCCAACTAAAATATGTATCGGCTGAAACTACGTTAAAAGAATCAGAAAAACAGGTCAAAGAAATAGAACAATTGAACAAACAGTTATCAGAAACTAAAAACAAAAACGAACTACTGCAACACGAAATAGATGCTTTAAAGGATGCATTAAGGCAAATGAAACATGATCCCTCATCGAACGTCGATGTGGCTTCACTTATCGACCACGCTGAAATCAAAGCCCAACTATCCAAAGCTTTAGTAGAAAGAAATAATTTAGCCAACAAGTATGAGGTCGAACAAAAGGAACGGGTTCGTCTCGATCAAGAACTGAAAAAAACTCAAGAGTCATTACATGCGACGAACCAGCAGGCTACAGAAGCCGTCACTAGACTTGAAGTTCTAGGAAAGTATTTCCAAGATCGGGAGAGCGAACTACTCAAGGAATTGAGTACTAAGGAATCATTGTGGCTGAGTAAACAGGGAGAATCTGCTAGCACCGTTGAGAAAATTGAACTACTGCAGCAAGAACTACAGAGATTCAAAGAGAAGTGCGACACGCTGAGCCGCGAGCTGGCGGAGCAGGAGTCGCTGCGGCGCGCGGCGGTGGGGGAGGTGGAGGCGCGCGCGCACACGGCGTGGCTGGAGGCGCGCGCCGCCCGCCGCGACGCCGACGCCGCGCGGGACCTCGCCGCCGCGCTGCGCCGCAAGCTCGCCGCCACCGCGCACGATGGCGCCCTCTCGCCGCATCACAAAGTAGCGTCACCTCTGGAAATGAGCGAGAACCCGGTGTTGCTGCCGCCGCCGCTGCCGCCTTTGTCGTTCCTACCGCCGCCGCTGTTACCGCCCCTGCCGAGACCGCCTCCACTCGGCAGGCTACCCTCGCCTCTCCAACCTAGGTACGGGGATCGACGCTACTCGCCGGACTCGCGGTACTCGCCGCAGAGGCGGTACTCGCCCGAGAGCCGCTACTCGCCGGAGACGGTGCGGTACTCGCCCGACAGCCGCTACTCGCCGCCGCCTCGCCGGTCGCCCTACTCGCCCAGACCCAGCAGACGACGTTCCAGAGATAGATACGAAGGTAGTTGTGGATCAAGGTCTCGTAACGCGCCCGAAACAGAATCCGAATACCCTTCGGACAGTCCATCAAGGAGCACGCGGCGACGGAGCAGATACCAGAGGCATTCAGGTCCCAGCTCTGGCTCAGGTTGTTCCTCGGAATCTGATAAATAA
Protein
MTVIKFILVLIFIAPSYGRVPDKVYCLDETCSKPLSRAKTLITYSSADPDHISYRNNAEALVFMKSAGTNPELWFASIEVPTESLIPKISFQPDKVKQPHEVVEGTTLYTTEATVKDINTAPQPASSLPNEINENLGTIKPVDNINPKNVDFENIREVNKRLLDTPNFPQQKIPNVQEAQENIQKKNTIETNVRFENVPNQHLNVHKDVNVHNPETKDQNYYKQVNLNEAELQKTSETLTTVPIQKFNEENFHDHNILNEKSVQKGHEQHVNDLPIIENSSPNVHNKEVYNEPQISISQSSSRSGSEGNIPNTEGIIHQMLNDEVPESEQNVSVPSNMNRNEDSDKHAKQAVADSTVENTQQQNNVYNNNQKGHEQHVNDLPIIENSSPNIHNKEVYNEPQISISQSSSRSGSEGNIPNTEGTIHQMLNDEVPESEQNVSVPSNMNRNEDRDKHAKQAVADSTVENTQQQNNLYNNIPNINGEQVQKNPELPLETNREQNNFLDQQNHPNEKLNKNHFEKSTPFMENPSNILYDNTQSEINRLTETISTKTEIESERTLPIVNRSPNSDNVVNYETGIPSTLSVTERNIEINNPNTGMPPIVENPNNIIPNINLKEIQLESREVPSVDRSHSPEYVLNNQNENIRTPPHPAETQRYFETNNAMTEIPPVVENVNNIVHNANLKEQQLESIPPYVDRAPNQENVFVDVAEQLRTPPPPYLNPITENYQQDHFENPTTETSPLLFNPSTSAIPDISINEEQPLTTELNIQTTTEGSKEPFSGMDENTNINQELQSNQEVGDKHSYYTETPVDIEYGNTAESNLPTSDEYQVFPEETTKGWFASIFDTLAGMWTSSSDDQDISDNTEDSIYDEPKTDNAEEFSFIKYLIHSYYSVMGIDEETRVLFTSAGDACYTENYCDGQSNTNKNRLLTFLLTTASSVLLFTLGYYYIDIRRQDGRLIGTINSLQRDLLFTTKECEILKEELSTTKTKLAGIEDNSFGMDDMVQSLKEEINEMKAQNDRLRHSLDDNEKLLRVSENTAEELQNTLSEVENTLSELLAERANLEEQIAEINGKAQAFEEELITVSRDRDNFQLKYVSAETTLKESEKQVKEIEQLNKQLSETKNKNELLQHEIDALKDALRQMKHDPSSNVDVASLIDHAEIKAQLSKALVERNNLANKYEVEQKERVRLDQELKKTQESLHATNQQATEAVTRLEVLGKYFQDRESELLKELSTKESLWLSKQGESASTVEKIELLQQELQRFKEKCDTLSRELAEQESLRRAAVGEVEARAHTAWLEARAARRDADAARDLAAALRRKLAATAHDGALSPHHKVASPLEMSENPVLLPPPLPPLSFLPPPLLPPLPRPPPLGRLPSPLQPRYGDRRYSPDSRYSPQRRYSPESRYSPETVRYSPDSRYSPPPRRSPYSPRPSRRRSRDRYEGSCGSRSRNAPETESEYPSDSPSRSTRRRSRYQRHSGPSSGSGCSSESDK
Summary
Uniprot
Pubmed
EMBL
PRIDE
ProteinModelPortal
PDB
6FSA
E-value=1.86264e-06,
Score=129
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.993341
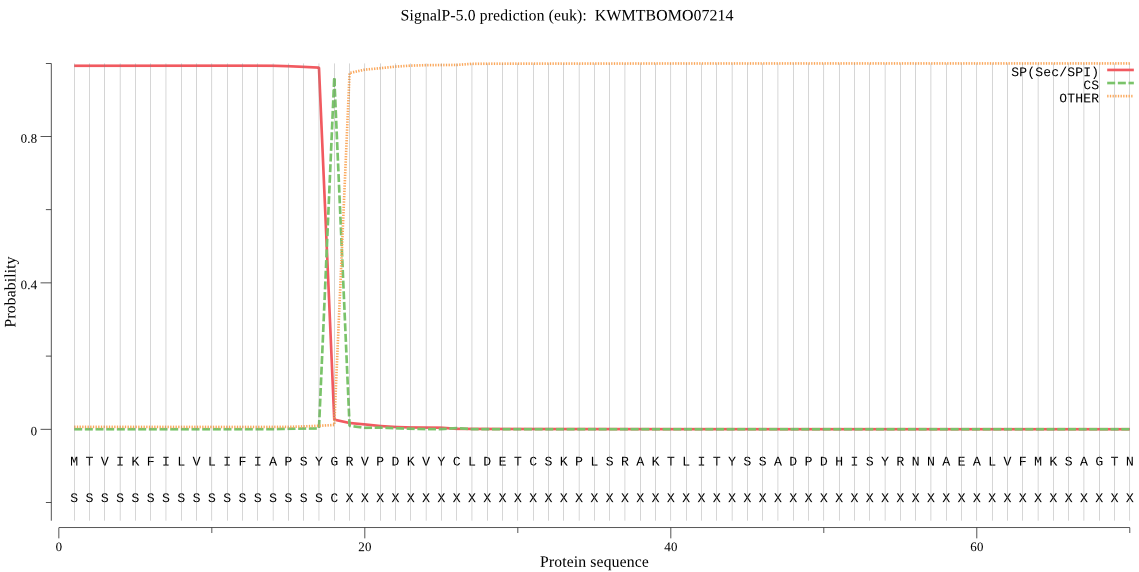
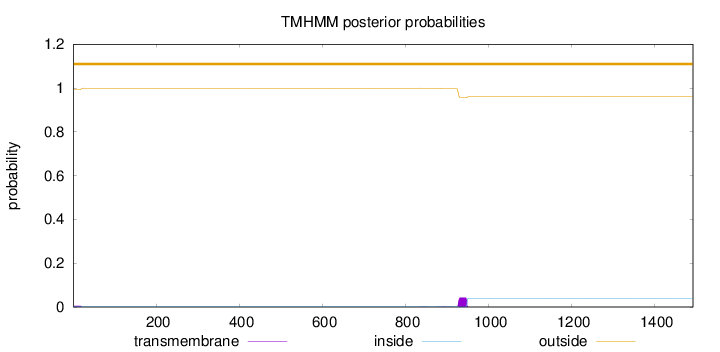
Length:
1492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.05476
Exp number, first 60 AAs:
0.10036
Total prob of N-in:
0.00609
outside
1 - 1492
Population Genetic Test Statistics
Pi
23.126904
Theta
20.15301
Tajima's D
-1.52293
CLR
142.827819
CSRT
0.0577471126443678
Interpretation
Uncertain
