Gene
KWMTBOMO07208
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.183 Nuclear Reliability : 1.536
Sequence
CDS
ATGCCACATGCCATCCGACTTTGGAATGAGCTTCCCTCCACGGTGTTTCCCGAGCGCTACGACATGTCCTTCTTCAAACAAGATTTGTGGACAGTACTTAACGGTAGGCAGCGGACGTCGCAGCGACGTGCAGATAGCAACGTCGTCTGCGGATTCATCTATGCAGACGACGTTGCTATCTGCACCGAGAGCCGCACAGAGCTACGAGAGGCACTCCTGTTGTGGAAACAGCAGTTACAAGCCGGTGGCCTAATATTGAGTGTTGCGAAGACACATTACATGTCTTTTAATGACCCAGATCCAGGTGACAACAGTCCGATTTCCATTGACGGTCAACTTGTAAACATGTGCGATCAATACAAGTATCTGGGTACTATGATGCACAGATCTGGAAATTTAAAATGCAACATTCAACACCGAATAGCCGCAGCGTGGCTAAAATGGCGAGAAGTAAGTGGTGTCACCTGCGATAGGAGGATGCCCGTTAAGCTCAAGGGTATGATCTATAAGACTATAATAAGGCCTGTTCTTATGTATGGCAGCGAAACATGGGCGGCAACCAAAAGGAACGTACATACCATTCAAGTTGCTGAAATGAAAATGCTGAGGTGGATGTGCGGCGTGACTAGGCTCGATAAAATCCGCAACGAGTACGTGCGTGGAAGTCTAGGTGTACGGGACATCGCCGACAAGATGCAGGAGAGTAGGCTGAGATGGTATGGTCACGTGAAAAGGAAGCCACCTGACTACGTCGGAAATTTGGCGCTACTGCTCGACCTCCCCGGTCGAAGACCTAGAGGCAGACCCAAGACAAGGTGGAGGGACGTAGTGCTGAAGGACAAGAGAGAATGCAATGCTGCTGACGAGGATGTCGAAGACAGAGCGAAGTGGAGGAGTAAAGTCAGGAAAGCTGACCCCACCACCATGTGGGATAAATAG
Protein
MPHAIRLWNELPSTVFPERYDMSFFKQDLWTVLNGRQRTSQRRADSNVVCGFIYADDVAICTESRTELREALLLWKQQLQAGGLILSVAKTHYMSFNDPDPGDNSPISIDGQLVNMCDQYKYLGTMMHRSGNLKCNIQHRIAAAWLKWREVSGVTCDRRMPVKLKGMIYKTIIRPVLMYGSETWAATKRNVHTIQVAEMKMLRWMCGVTRLDKIRNEYVRGSLGVRDIADKMQESRLRWYGHVKRKPPDYVGNLALLLDLPGRRPRGRPKTRWRDVVLKDKRECNAADEDVEDRAKWRSKVRKADPTTMWDK
Summary
Uniprot
D7F169
C6Y4D5
A0A2T8I3I2
A0A3B3HQB6
A0A3B3HJS0
A0A2T7D601
+ More
A0A2T8I3A3 A0A2S3IS22 A0A2S3HUX6 A0A2T8KNP5 A0A2T8KX40 A0A2T7C0N1 A0A2T8JCL0 A0A2S3HU98 Q1AKH8 A0A2T7C422 A0A2T7CCN9 I7K8L3 A0A3P9JDN6 A0A2T7F8M8 A0A2T8JCD8 A0A2T8KII0 A0A3Q1K9G2 A0A3P9JAC7 A0A3P9LFN7 A0A2T8I2Z1 A0A3P9I3Z4 A0A3P9MAU2 A0A3P9K1J1 A0A3B3H8Q9 A0A3P9IJG8 A0A3B3HJN2 A0A2T8HZM0 A0A3B3H6E8 A0A3Q1HIZ5 A0A3B3HFD6 A0A016UE98 A0A2T7F0C3 A0A3P9L6C8 A0A3P9HQ45 A0A2T8KVU2 A0A3B3I2L1 A0A3B3HGK1 A0A3B3H2V1 A0A1D6EFS5 A0A3B3HQ23 A0A3B3HVS1 A0A3P9IMA9 A0A3P9MDZ4 A0A3P9MJS1 A0A3B3H692 A0A3B3HIQ7 A0A3B3HSD0 A0A3B3ILX4 A0A3B3HTF4 A0A3B3HMM1 A0A3B3H644 A0A3B3IBX6 A0A3P9JLZ1 A0A3B3IM62 A0A3B3IDD8 A0A3B3H9R1 A0A3B3IM16 A0A3P9HCZ5 A0A3B3H5C8 A0A3B3IJY6 A0A3B3IEJ9 A0A3B3HVG5 A0A3B3IN57 A0A2T8I0K1 A0A2I0BDY2 A0A3B3HQS3 A0A3B3H879 A0A3B3H5B4 A0A3B3IIV8 A0A3B3HW87 A0A3P9MCH9 A0A2T7DY88 A0A3B3I4X3 A0A3P9HH58 A0A3B3I0B4 A0A3B3I8U4 A0A3B3HQW9 A0A3B3HYL2 A0A3P9IV37 A0A3B3H3Q1 A0A3B3INX9 A0A3B3IFB5 A0A3B3HXS2 A0A3B3ICX9 A0A3B3IC09 A0A3B3IEV4 A0A3B3HQP1 A0A3B3HT45 A0A3B3HV05 A0A3B3HQ01 A0A3P9JZH0 A0A2T7C6D7 A0A3P9KEZ0 A0A3P9LQU2
A0A2T8I3A3 A0A2S3IS22 A0A2S3HUX6 A0A2T8KNP5 A0A2T8KX40 A0A2T7C0N1 A0A2T8JCL0 A0A2S3HU98 Q1AKH8 A0A2T7C422 A0A2T7CCN9 I7K8L3 A0A3P9JDN6 A0A2T7F8M8 A0A2T8JCD8 A0A2T8KII0 A0A3Q1K9G2 A0A3P9JAC7 A0A3P9LFN7 A0A2T8I2Z1 A0A3P9I3Z4 A0A3P9MAU2 A0A3P9K1J1 A0A3B3H8Q9 A0A3P9IJG8 A0A3B3HJN2 A0A2T8HZM0 A0A3B3H6E8 A0A3Q1HIZ5 A0A3B3HFD6 A0A016UE98 A0A2T7F0C3 A0A3P9L6C8 A0A3P9HQ45 A0A2T8KVU2 A0A3B3I2L1 A0A3B3HGK1 A0A3B3H2V1 A0A1D6EFS5 A0A3B3HQ23 A0A3B3HVS1 A0A3P9IMA9 A0A3P9MDZ4 A0A3P9MJS1 A0A3B3H692 A0A3B3HIQ7 A0A3B3HSD0 A0A3B3ILX4 A0A3B3HTF4 A0A3B3HMM1 A0A3B3H644 A0A3B3IBX6 A0A3P9JLZ1 A0A3B3IM62 A0A3B3IDD8 A0A3B3H9R1 A0A3B3IM16 A0A3P9HCZ5 A0A3B3H5C8 A0A3B3IJY6 A0A3B3IEJ9 A0A3B3HVG5 A0A3B3IN57 A0A2T8I0K1 A0A2I0BDY2 A0A3B3HQS3 A0A3B3H879 A0A3B3H5B4 A0A3B3IIV8 A0A3B3HW87 A0A3P9MCH9 A0A2T7DY88 A0A3B3I4X3 A0A3P9HH58 A0A3B3I0B4 A0A3B3I8U4 A0A3B3HQW9 A0A3B3HYL2 A0A3P9IV37 A0A3B3H3Q1 A0A3B3INX9 A0A3B3IFB5 A0A3B3HXS2 A0A3B3ICX9 A0A3B3IC09 A0A3B3IEV4 A0A3B3HQP1 A0A3B3HT45 A0A3B3HV05 A0A3B3HQ01 A0A3P9JZH0 A0A2T7C6D7 A0A3P9KEZ0 A0A3P9LQU2
EMBL
FJ265554
ADI61822.1
CU462842
CBA11992.1
CM008054
PVH32202.1
+ More
CM009754 PUZ51002.1 PVH32156.1 PAN50441.2 CM008050 PAN30387.1 CM008047 PVH63795.1 CM008049 CM008046 PVH47980.1 PVH66734.1 CM009757 PUZ36875.1 PVH47648.1 PAN29912.1 DQ070849 ABB00038.1 PUZ38090.1 PUZ41102.1 HE805490 CCH50976.1 CM009749 PUZ76420.1 PVH47587.1 CM008048 PVH61993.1 PVH32022.1 PVH30886.1 JARK01001380 EYC13251.1 CM009750 PUZ73532.1 PVH66296.1 CM007648 ONM19028.1 PVH31207.1 KZ451888 PKA66013.1 CM009752 PUZ60542.1 PUZ38902.1
CM009754 PUZ51002.1 PVH32156.1 PAN50441.2 CM008050 PAN30387.1 CM008047 PVH63795.1 CM008049 CM008046 PVH47980.1 PVH66734.1 CM009757 PUZ36875.1 PVH47648.1 PAN29912.1 DQ070849 ABB00038.1 PUZ38090.1 PUZ41102.1 HE805490 CCH50976.1 CM009749 PUZ76420.1 PVH47587.1 CM008048 PVH61993.1 PVH32022.1 PVH30886.1 JARK01001380 EYC13251.1 CM009750 PUZ73532.1 PVH66296.1 CM007648 ONM19028.1 PVH31207.1 KZ451888 PKA66013.1 CM009752 PUZ60542.1 PUZ38902.1
Proteomes
PRIDE
Pfam
PF00078 RVT_1
Interpro
Gene 3D
ProteinModelPortal
D7F169
C6Y4D5
A0A2T8I3I2
A0A3B3HQB6
A0A3B3HJS0
A0A2T7D601
+ More
A0A2T8I3A3 A0A2S3IS22 A0A2S3HUX6 A0A2T8KNP5 A0A2T8KX40 A0A2T7C0N1 A0A2T8JCL0 A0A2S3HU98 Q1AKH8 A0A2T7C422 A0A2T7CCN9 I7K8L3 A0A3P9JDN6 A0A2T7F8M8 A0A2T8JCD8 A0A2T8KII0 A0A3Q1K9G2 A0A3P9JAC7 A0A3P9LFN7 A0A2T8I2Z1 A0A3P9I3Z4 A0A3P9MAU2 A0A3P9K1J1 A0A3B3H8Q9 A0A3P9IJG8 A0A3B3HJN2 A0A2T8HZM0 A0A3B3H6E8 A0A3Q1HIZ5 A0A3B3HFD6 A0A016UE98 A0A2T7F0C3 A0A3P9L6C8 A0A3P9HQ45 A0A2T8KVU2 A0A3B3I2L1 A0A3B3HGK1 A0A3B3H2V1 A0A1D6EFS5 A0A3B3HQ23 A0A3B3HVS1 A0A3P9IMA9 A0A3P9MDZ4 A0A3P9MJS1 A0A3B3H692 A0A3B3HIQ7 A0A3B3HSD0 A0A3B3ILX4 A0A3B3HTF4 A0A3B3HMM1 A0A3B3H644 A0A3B3IBX6 A0A3P9JLZ1 A0A3B3IM62 A0A3B3IDD8 A0A3B3H9R1 A0A3B3IM16 A0A3P9HCZ5 A0A3B3H5C8 A0A3B3IJY6 A0A3B3IEJ9 A0A3B3HVG5 A0A3B3IN57 A0A2T8I0K1 A0A2I0BDY2 A0A3B3HQS3 A0A3B3H879 A0A3B3H5B4 A0A3B3IIV8 A0A3B3HW87 A0A3P9MCH9 A0A2T7DY88 A0A3B3I4X3 A0A3P9HH58 A0A3B3I0B4 A0A3B3I8U4 A0A3B3HQW9 A0A3B3HYL2 A0A3P9IV37 A0A3B3H3Q1 A0A3B3INX9 A0A3B3IFB5 A0A3B3HXS2 A0A3B3ICX9 A0A3B3IC09 A0A3B3IEV4 A0A3B3HQP1 A0A3B3HT45 A0A3B3HV05 A0A3B3HQ01 A0A3P9JZH0 A0A2T7C6D7 A0A3P9KEZ0 A0A3P9LQU2
A0A2T8I3A3 A0A2S3IS22 A0A2S3HUX6 A0A2T8KNP5 A0A2T8KX40 A0A2T7C0N1 A0A2T8JCL0 A0A2S3HU98 Q1AKH8 A0A2T7C422 A0A2T7CCN9 I7K8L3 A0A3P9JDN6 A0A2T7F8M8 A0A2T8JCD8 A0A2T8KII0 A0A3Q1K9G2 A0A3P9JAC7 A0A3P9LFN7 A0A2T8I2Z1 A0A3P9I3Z4 A0A3P9MAU2 A0A3P9K1J1 A0A3B3H8Q9 A0A3P9IJG8 A0A3B3HJN2 A0A2T8HZM0 A0A3B3H6E8 A0A3Q1HIZ5 A0A3B3HFD6 A0A016UE98 A0A2T7F0C3 A0A3P9L6C8 A0A3P9HQ45 A0A2T8KVU2 A0A3B3I2L1 A0A3B3HGK1 A0A3B3H2V1 A0A1D6EFS5 A0A3B3HQ23 A0A3B3HVS1 A0A3P9IMA9 A0A3P9MDZ4 A0A3P9MJS1 A0A3B3H692 A0A3B3HIQ7 A0A3B3HSD0 A0A3B3ILX4 A0A3B3HTF4 A0A3B3HMM1 A0A3B3H644 A0A3B3IBX6 A0A3P9JLZ1 A0A3B3IM62 A0A3B3IDD8 A0A3B3H9R1 A0A3B3IM16 A0A3P9HCZ5 A0A3B3H5C8 A0A3B3IJY6 A0A3B3IEJ9 A0A3B3HVG5 A0A3B3IN57 A0A2T8I0K1 A0A2I0BDY2 A0A3B3HQS3 A0A3B3H879 A0A3B3H5B4 A0A3B3IIV8 A0A3B3HW87 A0A3P9MCH9 A0A2T7DY88 A0A3B3I4X3 A0A3P9HH58 A0A3B3I0B4 A0A3B3I8U4 A0A3B3HQW9 A0A3B3HYL2 A0A3P9IV37 A0A3B3H3Q1 A0A3B3INX9 A0A3B3IFB5 A0A3B3HXS2 A0A3B3ICX9 A0A3B3IC09 A0A3B3IEV4 A0A3B3HQP1 A0A3B3HT45 A0A3B3HV05 A0A3B3HQ01 A0A3P9JZH0 A0A2T7C6D7 A0A3P9KEZ0 A0A3P9LQU2
Ontologies
KEGG
PANTHER
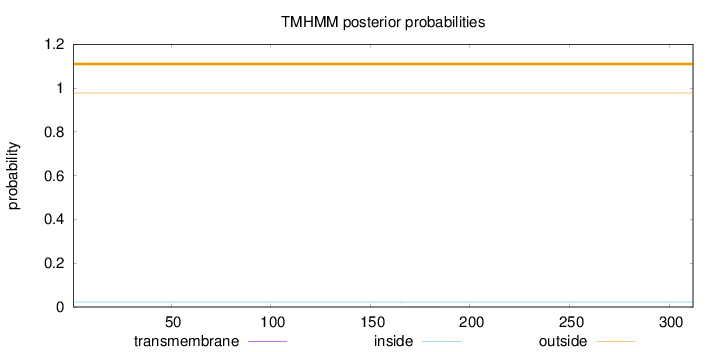
Topology
Length:
312
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00531
Exp number, first 60 AAs:
0.00105
Total prob of N-in:
0.02261
outside
1 - 312
Population Genetic Test Statistics
Pi
273.518458
Theta
150.232955
Tajima's D
4.547944
CLR
0
CSRT
0.999950002499875
Interpretation
Uncertain
