Gene
KWMTBOMO07206
Pre Gene Modal
BGIBMGA010573
Annotation
PREDICTED:_uncharacterized_protein_LOC101746257_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.995
Sequence
CDS
ATGTCGACTCGCGCGTACGTAGCGCGATCAAGAACTATGACCAGTTTGCGAGTACTCCTTGTTTGTGTGCTTTTAATCAACGGTACACATCAACAGAAAGATAAGGAGGAAGAGCAGTTTGTTTGTCCCGAAGGAACACAGGGTAATGGGAATTTCGCCGACCCAGCCACCTGCAGACGTTTTTATCAATGTGTCGACGGGTACCCGTATTTAAACAGATGTCCTTCAGGATTATATTTTGATGACATCAGCAAATACTGCACCTTCAAAGCAGAAGCGAGGTGTGGACCCATACCAACTACACCGGCGCCTATAACAGAAGTACCGCTCGATTTAGCTACAAAGTGCGAGCCTGCGGAATGCCAATTACCGTACTGCTTCTGTTCCAAAGATGGTACATTGATTCCTGGCGGCCTCGACCCTGCAGAAACACCTCAAATGATAATGTTGACTTTCGATGGAGCCGTAAATTTGAACAACTTCGACAAGTACAAGAAAGTCTTCAAAGGAAAGATTAACAATCCAAACGGATGTCCTATAAAGGGGACATTCTTCTTGTCTCATGAATACAGTAACTACGTGATGGTACAAGCTCTCGCACACGATGGACATGAGATAGCAACAGGTACAGTTTCGCAACAGCAAGGCTTGCAAGACAAAGGGTACGAGGAATGGGCCGGTGAGATGATCGGCATGCGAGAGATCCTTAAGAAGTTTGCGAACATATCGCGGTCGGAGATCGTGGGAGCCCGAGCACCGTTTCTGAAGCCTGGCAGGAATACGCAGTTCAAAGTGTTAGAAGACTTCGGATACATCTACGACAGTTCAGTGGGCGTGCCTCCTTTGCCGATTCCGGTGTGGCCTTACACGCTCGACTACAAGATCGCCCACGAGTGCAAGTCCGGAACATGCCCCACTAAATCGTTCCCGGGACTTTGGGAGATACCATTCAATGCTCACTACGTGGAATCATTCGAGGGAGGTCATTGTCCTTACTTGGACCAATGCGTTCTACATAATCACGATGCTAACGAGGTATTCGAATGGTTACAAGAAGACTTCAGTCGTTATTACGAGCAGAACAGAGCCCCATACATGATGCCGCTTCACACCAATTGGTTCCAAATCAAAGAACTTGAGCTTGGTCTACATAAGTTCTTGAAATGGGCGGCAGATCTGGAAGACGTATGGTTTGTAACAATGACACAAAGTTTAACATGGATGACGGATCCACGACCAGTGAAGGCGCTCAACAACTACGAAGCGTGGCGTTGTGACAACAAAGAGCTGCCTTCCGCTCCCTGCAACTTGCCTAATAAGTGTGCTCTTTCGTTCAAACAACCGGATAGCAATTTCACTGATACAAGGTACATGGAGACTTGCAGCGAGTGCCCTAACCAGTACCCGTGGCTAGGTGATTCTGGTGGATCCGGAATACCCGGTAAGGATAATTATATACCCGAGAACCTTAAGAGGAAGTAG
Protein
MSTRAYVARSRTMTSLRVLLVCVLLINGTHQQKDKEEEQFVCPEGTQGNGNFADPATCRRFYQCVDGYPYLNRCPSGLYFDDISKYCTFKAEARCGPIPTTPAPITEVPLDLATKCEPAECQLPYCFCSKDGTLIPGGLDPAETPQMIMLTFDGAVNLNNFDKYKKVFKGKINNPNGCPIKGTFFLSHEYSNYVMVQALAHDGHEIATGTVSQQQGLQDKGYEEWAGEMIGMREILKKFANISRSEIVGARAPFLKPGRNTQFKVLEDFGYIYDSSVGVPPLPIPVWPYTLDYKIAHECKSGTCPTKSFPGLWEIPFNAHYVESFEGGHCPYLDQCVLHNHDANEVFEWLQEDFSRYYEQNRAPYMMPLHTNWFQIKELELGLHKFLKWAADLEDVWFVTMTQSLTWMTDPRPVKALNNYEAWRCDNKELPSAPCNLPNKCALSFKQPDSNFTDTRYMETCSECPNQYPWLGDSGGSGIPGKDNYIPENLKRK
Summary
Uniprot
A0A2H1W7A7
H9JM21
A0A0G2RLM8
A0A194PJ81
A0A3S2P1L0
A0A194R5P1
+ More
A0A2Z5U8C0 A0A212FL49 A0A2A4ITV9 A8W489 A0A2Z4N549 A0A2R7W7I6 A0A2A3EFF5 A0A0F6PK41 A0A088A256 A0A1B6BYV2 A0A146KML0 A0A0A9ZAW6 A0A1W4WHB3 E2AIL9 U4UP63 F4W6Y8 A0A067RMF9 A0A182IAZ5 K7IUS9 Q7PS06 A0A182WTJ3 A0A0J7NTK4 E2BZS8 A0A182R281 T1HG33 A0A195EWC1 A0A182W657 A0A182ULW4 A0A1B6J002 A0A151HZB8 A0A084VFS9 A0A195DUZ0 A0A182YF09 E0VP39 A0A158NSM4 A0A195CQE3 B0WH33 A0A1B6GEZ7 A0A026WS59 N6TQX2 A0A3L8D7J5 N6TG25 N6UCF7 A0A1Y1MJY7 A0A0L7R5V5 A0A310SAB3 Q179T5 A0A182Q9J6 A0A1Q3FMN8 A0A2S2Q5M5 A0A232F0R5 J9JRK2 A0A182N0Q6 A0A1J1J1J1 T1PGI6 W8BPN2 A0A0L0BWH4 E9IDK3 A0A336K435 B4JLI9 A0A182U775 A0A0A1WUL6 A0A1I8Q7E9 B4MST9 B4L3X5 B4PZL3 B4ILE6 B3NY80 A0A3B0K0T5 Q9VR69 B4M3L2 A0A3B0JDE5 B3MYK7 A0A0K8TZY1 A0A0P6JZ94 A0A1W4VYP5 A0A2Z2Z006 A0A034WUN8 A0A2S2NGL5 A0A1S3DFS2 A0A182J9N2 A0A182FIM5 W5JHB7 A0A1B6MGC0 A0A226E5M1 A0A1D2N0D8 A0A1B0DM47 A0A0N0BHK4 A0A2J7Q891 A0A182LWD8 A0A1B0CP06 A0A3Q0IJU1 A0A224XFM2 A0A1B0FGZ2
A0A2Z5U8C0 A0A212FL49 A0A2A4ITV9 A8W489 A0A2Z4N549 A0A2R7W7I6 A0A2A3EFF5 A0A0F6PK41 A0A088A256 A0A1B6BYV2 A0A146KML0 A0A0A9ZAW6 A0A1W4WHB3 E2AIL9 U4UP63 F4W6Y8 A0A067RMF9 A0A182IAZ5 K7IUS9 Q7PS06 A0A182WTJ3 A0A0J7NTK4 E2BZS8 A0A182R281 T1HG33 A0A195EWC1 A0A182W657 A0A182ULW4 A0A1B6J002 A0A151HZB8 A0A084VFS9 A0A195DUZ0 A0A182YF09 E0VP39 A0A158NSM4 A0A195CQE3 B0WH33 A0A1B6GEZ7 A0A026WS59 N6TQX2 A0A3L8D7J5 N6TG25 N6UCF7 A0A1Y1MJY7 A0A0L7R5V5 A0A310SAB3 Q179T5 A0A182Q9J6 A0A1Q3FMN8 A0A2S2Q5M5 A0A232F0R5 J9JRK2 A0A182N0Q6 A0A1J1J1J1 T1PGI6 W8BPN2 A0A0L0BWH4 E9IDK3 A0A336K435 B4JLI9 A0A182U775 A0A0A1WUL6 A0A1I8Q7E9 B4MST9 B4L3X5 B4PZL3 B4ILE6 B3NY80 A0A3B0K0T5 Q9VR69 B4M3L2 A0A3B0JDE5 B3MYK7 A0A0K8TZY1 A0A0P6JZ94 A0A1W4VYP5 A0A2Z2Z006 A0A034WUN8 A0A2S2NGL5 A0A1S3DFS2 A0A182J9N2 A0A182FIM5 W5JHB7 A0A1B6MGC0 A0A226E5M1 A0A1D2N0D8 A0A1B0DM47 A0A0N0BHK4 A0A2J7Q891 A0A182LWD8 A0A1B0CP06 A0A3Q0IJU1 A0A224XFM2 A0A1B0FGZ2
Pubmed
19121390
26354079
22118469
18362917
19820115
24989071
+ More
26823975 25401762 20798317 23537049 21719571 24845553 20075255 12364791 24438588 25244985 20566863 21347285 24508170 30249741 28004739 17510324 28648823 25315136 24495485 26108605 21282665 17994087 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 20920257 23761445 27289101
26823975 25401762 20798317 23537049 21719571 24845553 20075255 12364791 24438588 25244985 20566863 21347285 24508170 30249741 28004739 17510324 28648823 25315136 24495485 26108605 21282665 17994087 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 20920257 23761445 27289101
EMBL
ODYU01006772
SOQ48923.1
BABH01005714
BABH01005715
BABH01005716
BABH01005717
+ More
KP000853 AJG44548.1 KQ459603 KPI92779.1 RSAL01000003 RVE54759.1 KQ460883 KPJ11181.1 AP017552 BBB06981.1 AGBW02007871 OWR54464.1 NWSH01007543 PCG62926.1 EU190486 KQ971338 ABW74146.1 EFA02013.1 MF612053 AWX65385.1 KK854405 PTY15539.1 KZ288260 PBC30467.1 KJ825889 AJQ20734.1 GEDC01031133 JAS06165.1 GDHC01020835 JAP97793.1 GBHO01003111 JAG40493.1 GL439840 EFN66708.1 KB632308 ERL91941.1 GL887793 EGI70042.1 KK852424 KDR24193.1 APCN01001396 AAZX01005919 AAAB01008846 EAA06323.5 LBMM01001782 KMQ95755.1 GL451666 EFN78807.1 ACPB03001124 KQ981948 KYN32575.1 GECU01015220 JAS92486.1 KQ976707 KYM77066.1 ATLV01012459 KE524793 KFB36823.1 KQ980322 KYN16651.1 DS235354 EEB15145.1 ADTU01025008 KQ977444 KYN02697.1 DS231932 EDS27484.1 GECZ01008744 JAS61025.1 KK107111 EZA58885.1 APGK01009027 APGK01009028 APGK01009029 KB737836 ENN82894.1 QOIP01000011 RLU16455.1 APGK01029136 APGK01029137 APGK01029138 APGK01029139 KB740725 ENN79344.1 ENN79345.1 GEZM01029622 GEZM01029621 GEZM01029620 GEZM01029619 JAV86041.1 KQ414648 KOC66272.1 KQ762237 OAD56001.1 CH477346 EAT42983.1 AXCN02001397 GFDL01006267 JAV28778.1 GGMS01003834 MBY73037.1 NNAY01001398 OXU24113.1 ABLF02037278 CVRI01000063 CRL04729.1 KA647255 AFP61884.1 GAMC01003395 JAC03161.1 JRES01001325 KNC23614.1 GL762454 EFZ21332.1 UFQS01000017 UFQT01000017 SSW97473.1 SSX17859.1 CH916370 EDW00442.1 GBXI01011548 JAD02744.1 CH963851 EDW75178.1 CH933810 EDW07253.2 CM000162 EDX03141.1 CH480870 EDW53805.1 CH954180 EDV47665.1 OUUW01000003 SPP78631.1 AE014298 AY113493 AAF50937.2 AAM29498.1 CH940651 EDW65387.1 SPP78633.1 CH902632 EDV32701.2 GDHF01032491 JAI19823.1 GDIQ01002178 JAN92559.1 KY681051 AUP42578.1 GAKP01000900 JAC58052.1 GGMR01003453 MBY16072.1 ADMH02001198 ETN63762.1 GEBQ01004979 JAT34998.1 LNIX01000006 OXA52729.1 LJIJ01000328 ODM98739.1 AJVK01016665 AJVK01016666 KQ435750 KOX76384.1 NEVH01016966 PNF24804.1 AXCM01007884 AJWK01021258 AJWK01021259 AJWK01021260 AJWK01021261 AJWK01021262 AJWK01021263 GFTR01006592 JAW09834.1 CCAG010009005 CCAG010009006
KP000853 AJG44548.1 KQ459603 KPI92779.1 RSAL01000003 RVE54759.1 KQ460883 KPJ11181.1 AP017552 BBB06981.1 AGBW02007871 OWR54464.1 NWSH01007543 PCG62926.1 EU190486 KQ971338 ABW74146.1 EFA02013.1 MF612053 AWX65385.1 KK854405 PTY15539.1 KZ288260 PBC30467.1 KJ825889 AJQ20734.1 GEDC01031133 JAS06165.1 GDHC01020835 JAP97793.1 GBHO01003111 JAG40493.1 GL439840 EFN66708.1 KB632308 ERL91941.1 GL887793 EGI70042.1 KK852424 KDR24193.1 APCN01001396 AAZX01005919 AAAB01008846 EAA06323.5 LBMM01001782 KMQ95755.1 GL451666 EFN78807.1 ACPB03001124 KQ981948 KYN32575.1 GECU01015220 JAS92486.1 KQ976707 KYM77066.1 ATLV01012459 KE524793 KFB36823.1 KQ980322 KYN16651.1 DS235354 EEB15145.1 ADTU01025008 KQ977444 KYN02697.1 DS231932 EDS27484.1 GECZ01008744 JAS61025.1 KK107111 EZA58885.1 APGK01009027 APGK01009028 APGK01009029 KB737836 ENN82894.1 QOIP01000011 RLU16455.1 APGK01029136 APGK01029137 APGK01029138 APGK01029139 KB740725 ENN79344.1 ENN79345.1 GEZM01029622 GEZM01029621 GEZM01029620 GEZM01029619 JAV86041.1 KQ414648 KOC66272.1 KQ762237 OAD56001.1 CH477346 EAT42983.1 AXCN02001397 GFDL01006267 JAV28778.1 GGMS01003834 MBY73037.1 NNAY01001398 OXU24113.1 ABLF02037278 CVRI01000063 CRL04729.1 KA647255 AFP61884.1 GAMC01003395 JAC03161.1 JRES01001325 KNC23614.1 GL762454 EFZ21332.1 UFQS01000017 UFQT01000017 SSW97473.1 SSX17859.1 CH916370 EDW00442.1 GBXI01011548 JAD02744.1 CH963851 EDW75178.1 CH933810 EDW07253.2 CM000162 EDX03141.1 CH480870 EDW53805.1 CH954180 EDV47665.1 OUUW01000003 SPP78631.1 AE014298 AY113493 AAF50937.2 AAM29498.1 CH940651 EDW65387.1 SPP78633.1 CH902632 EDV32701.2 GDHF01032491 JAI19823.1 GDIQ01002178 JAN92559.1 KY681051 AUP42578.1 GAKP01000900 JAC58052.1 GGMR01003453 MBY16072.1 ADMH02001198 ETN63762.1 GEBQ01004979 JAT34998.1 LNIX01000006 OXA52729.1 LJIJ01000328 ODM98739.1 AJVK01016665 AJVK01016666 KQ435750 KOX76384.1 NEVH01016966 PNF24804.1 AXCM01007884 AJWK01021258 AJWK01021259 AJWK01021260 AJWK01021261 AJWK01021262 AJWK01021263 GFTR01006592 JAW09834.1 CCAG010009005 CCAG010009006
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000007151
UP000218220
+ More
UP000007266 UP000242457 UP000005203 UP000192223 UP000000311 UP000030742 UP000007755 UP000027135 UP000075840 UP000002358 UP000007062 UP000076407 UP000036403 UP000008237 UP000075900 UP000015103 UP000078541 UP000075920 UP000075903 UP000078540 UP000030765 UP000078492 UP000076408 UP000009046 UP000005205 UP000078542 UP000002320 UP000053097 UP000019118 UP000279307 UP000053825 UP000008820 UP000075886 UP000215335 UP000007819 UP000075884 UP000183832 UP000095301 UP000037069 UP000001070 UP000075902 UP000095300 UP000007798 UP000009192 UP000002282 UP000001292 UP000008711 UP000268350 UP000000803 UP000008792 UP000007801 UP000192221 UP000079169 UP000075880 UP000069272 UP000000673 UP000198287 UP000094527 UP000092462 UP000053105 UP000235965 UP000075883 UP000092461 UP000092444
UP000007266 UP000242457 UP000005203 UP000192223 UP000000311 UP000030742 UP000007755 UP000027135 UP000075840 UP000002358 UP000007062 UP000076407 UP000036403 UP000008237 UP000075900 UP000015103 UP000078541 UP000075920 UP000075903 UP000078540 UP000030765 UP000078492 UP000076408 UP000009046 UP000005205 UP000078542 UP000002320 UP000053097 UP000019118 UP000279307 UP000053825 UP000008820 UP000075886 UP000215335 UP000007819 UP000075884 UP000183832 UP000095301 UP000037069 UP000001070 UP000075902 UP000095300 UP000007798 UP000009192 UP000002282 UP000001292 UP000008711 UP000268350 UP000000803 UP000008792 UP000007801 UP000192221 UP000079169 UP000075880 UP000069272 UP000000673 UP000198287 UP000094527 UP000092462 UP000053105 UP000235965 UP000075883 UP000092461 UP000092444
Interpro
ProteinModelPortal
A0A2H1W7A7
H9JM21
A0A0G2RLM8
A0A194PJ81
A0A3S2P1L0
A0A194R5P1
+ More
A0A2Z5U8C0 A0A212FL49 A0A2A4ITV9 A8W489 A0A2Z4N549 A0A2R7W7I6 A0A2A3EFF5 A0A0F6PK41 A0A088A256 A0A1B6BYV2 A0A146KML0 A0A0A9ZAW6 A0A1W4WHB3 E2AIL9 U4UP63 F4W6Y8 A0A067RMF9 A0A182IAZ5 K7IUS9 Q7PS06 A0A182WTJ3 A0A0J7NTK4 E2BZS8 A0A182R281 T1HG33 A0A195EWC1 A0A182W657 A0A182ULW4 A0A1B6J002 A0A151HZB8 A0A084VFS9 A0A195DUZ0 A0A182YF09 E0VP39 A0A158NSM4 A0A195CQE3 B0WH33 A0A1B6GEZ7 A0A026WS59 N6TQX2 A0A3L8D7J5 N6TG25 N6UCF7 A0A1Y1MJY7 A0A0L7R5V5 A0A310SAB3 Q179T5 A0A182Q9J6 A0A1Q3FMN8 A0A2S2Q5M5 A0A232F0R5 J9JRK2 A0A182N0Q6 A0A1J1J1J1 T1PGI6 W8BPN2 A0A0L0BWH4 E9IDK3 A0A336K435 B4JLI9 A0A182U775 A0A0A1WUL6 A0A1I8Q7E9 B4MST9 B4L3X5 B4PZL3 B4ILE6 B3NY80 A0A3B0K0T5 Q9VR69 B4M3L2 A0A3B0JDE5 B3MYK7 A0A0K8TZY1 A0A0P6JZ94 A0A1W4VYP5 A0A2Z2Z006 A0A034WUN8 A0A2S2NGL5 A0A1S3DFS2 A0A182J9N2 A0A182FIM5 W5JHB7 A0A1B6MGC0 A0A226E5M1 A0A1D2N0D8 A0A1B0DM47 A0A0N0BHK4 A0A2J7Q891 A0A182LWD8 A0A1B0CP06 A0A3Q0IJU1 A0A224XFM2 A0A1B0FGZ2
A0A2Z5U8C0 A0A212FL49 A0A2A4ITV9 A8W489 A0A2Z4N549 A0A2R7W7I6 A0A2A3EFF5 A0A0F6PK41 A0A088A256 A0A1B6BYV2 A0A146KML0 A0A0A9ZAW6 A0A1W4WHB3 E2AIL9 U4UP63 F4W6Y8 A0A067RMF9 A0A182IAZ5 K7IUS9 Q7PS06 A0A182WTJ3 A0A0J7NTK4 E2BZS8 A0A182R281 T1HG33 A0A195EWC1 A0A182W657 A0A182ULW4 A0A1B6J002 A0A151HZB8 A0A084VFS9 A0A195DUZ0 A0A182YF09 E0VP39 A0A158NSM4 A0A195CQE3 B0WH33 A0A1B6GEZ7 A0A026WS59 N6TQX2 A0A3L8D7J5 N6TG25 N6UCF7 A0A1Y1MJY7 A0A0L7R5V5 A0A310SAB3 Q179T5 A0A182Q9J6 A0A1Q3FMN8 A0A2S2Q5M5 A0A232F0R5 J9JRK2 A0A182N0Q6 A0A1J1J1J1 T1PGI6 W8BPN2 A0A0L0BWH4 E9IDK3 A0A336K435 B4JLI9 A0A182U775 A0A0A1WUL6 A0A1I8Q7E9 B4MST9 B4L3X5 B4PZL3 B4ILE6 B3NY80 A0A3B0K0T5 Q9VR69 B4M3L2 A0A3B0JDE5 B3MYK7 A0A0K8TZY1 A0A0P6JZ94 A0A1W4VYP5 A0A2Z2Z006 A0A034WUN8 A0A2S2NGL5 A0A1S3DFS2 A0A182J9N2 A0A182FIM5 W5JHB7 A0A1B6MGC0 A0A226E5M1 A0A1D2N0D8 A0A1B0DM47 A0A0N0BHK4 A0A2J7Q891 A0A182LWD8 A0A1B0CP06 A0A3Q0IJU1 A0A224XFM2 A0A1B0FGZ2
PDB
5ZNT
E-value=1.44029e-74,
Score=712
Ontologies
GO
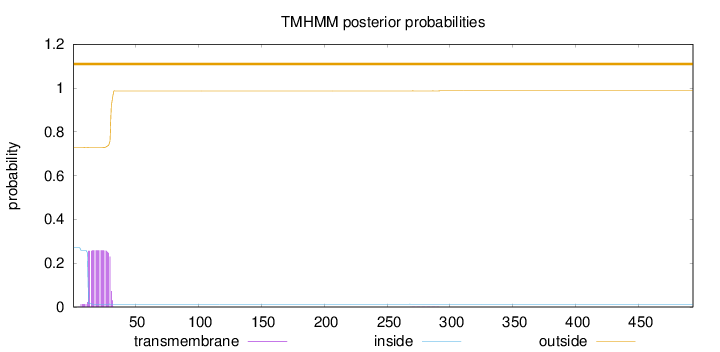
Topology
SignalP
Position: 1 - 31,
Likelihood: 0.960579
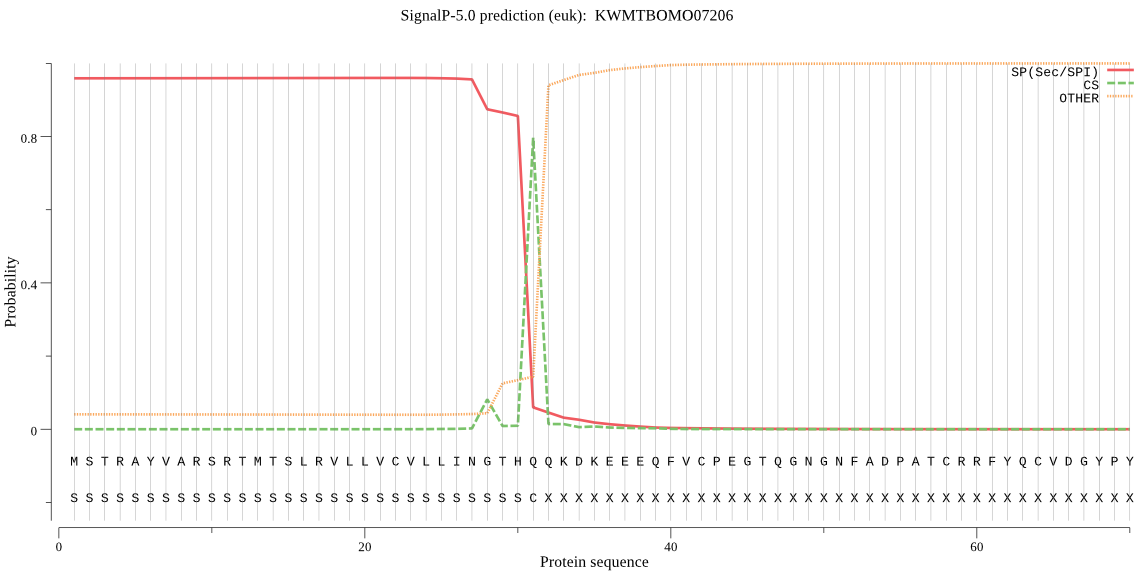
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.80582999999998
Exp number, first 60 AAs:
4.79195
Total prob of N-in:
0.27090
outside
1 - 493
Population Genetic Test Statistics
Pi
211.160054
Theta
163.526171
Tajima's D
-1.973009
CLR
0.257868
CSRT
0.0165491725413729
Interpretation
Possibly Positive selection
