Gene
KWMTBOMO07205 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010381
Annotation
PREDICTED:_protein_sly1_homolog_[Papilio_polytes]
Full name
Protein sly1 homolog
Location in the cell
Nuclear Reliability : 2.083
Sequence
CDS
ATGTCTACGCTAAGAGAACGCCAACTTAATGCCTTGAAACAGATGCTTAATTTAAATCAGCCGCTTACAAAGAATATAGCCAATGAACCGACGTGGAAAATCTTAATATATGATAGAGTTGGACAAGATATTATCTCACCATTGGTATCTGTAAAAGATTTGCGTGAACTAGGAGTGACTCTTCATTTACAACTACATTCAGATCGAGATTCGATACCAGAAGTACCTGCTGTTTATTTCTGTTCTCCTTCTGATGAAAATCTGGATCGCATTTGCCAAGATCTCGATAATGGGATCTATGATCAATACCATTTAAACTTTATATCACCAATCACAAGACACAAACTTGAAGACTTAGCTGCATCAGCTATCCAGTCAAATTCTGCTGTCAATATACATAAAGTTTTCGATCAGTATTTGAACTTTATTTGTTTGGAAGATGATCTTTTTATAATGAAGCACCAGCAATCTGATTCATTGTCATATTATGCAATAAACAAAGCTGATACTAAGGACACTGAAATGGAAGTGATCATGGACAACATTGTTGAAAGCCTGTTCTCTGTGTTTGTGACAATGGGCAATGTACCTTTTATACGAAGTACTAAAGGTAATGCTGCTGAGATGGTTGCTAAAAAGCTTGATAAAAAATTGAGAGAAAATCTTTGGGATGCCCGAAACAATCTCTTCCATGGCAACACTGGACAAGGTGGTGCATTCAGTTTTACGAGACCCATGTTGATTCTACTGGATAGAAATATTGACATGGCAACACCTCTCCATCACACATGGACCTATCAAGCACTTGCACATGATGTATTGGATTTATCACTGAATAGAGCAGTTGTCCCAGAAAACACAAATGCTACAGTACCGATTCAGAAATCAAACACGAGGATATGTGACCTTGATTCGAAAGATCCATTATGGTCAGAGCACAAAGGTAGCCCCTTTCCAACTGTTGCTGAGGCTATACAAGAGGATTTAGATAAATACAGGAGTTCAGAAGCCGAAGTAAAGAAACTGAAGAGCTCTATGGGTCTCGATGCAGAAAGTGACTTGGCCCTGAGCATGGTCAGTGACAATACTCAACGGCTTACCAACGCGGTGAACTCATTACCTCAACTCATGGAAAAGAAACGGCTGATTGATATGCACACTACTATCGCCACAGCAATTCTTAATGCCATTAAGTCCAGAAGACTAGATTCATTTTTCGAATTAGAGGAAAAAATAATGAGTAAAAGCACCGCCGTGGAAACCAAAGCGGTTATGGATCTTATAACTGATCCCAGTGCGGGTACACCCGAGGACAAAATGAGGCTGTTCATTATTTACTATCTGTGCACGTCGCAATTCACTGACGATGAATATGAAAAGTTTGAAGGAGCTCTAGTGGCGGCCAACTGCGATGTCAAAGCCCTATCTTACATGAAACGGTGGAAGGGTTTCACAAAAATGTCAAATCAGTACGAAGGCGGCGGCACCAAAACAGTATCCATGTTCTCTAAACTTGTATCCCAAGGCTCATCCTTTGTAATGGAGGGCGTCAAAAATTTAGTTGTCAAAAAACACAAACTTCCCGTGAGCCGCATAGTGGAGAGCGCTCTGAGTGGCAGCGAGAGCGCCGCCGAGCTCACGTGGCTGGACCCCCGCGCGGCGCGCACGGACTCGGCCGGTCGCGCGCGCTCCGCACGACAACAGCCGCCCACCGACGCTGTGGTCTTCCTCGTCGGCGGCGGGAATTACATCGAATACCACAACCTGATCGACTTCGCTAAGCAACAAGCAGCAAATGGCATACCTAGAAAACTGATTTATGGTGCTACAACTTTGCCAAACGGGTCCCAATTTTTAAAACAGTTGTCGCTACTCGGAGAAGAGATTCAATGA
Protein
MSTLRERQLNALKQMLNLNQPLTKNIANEPTWKILIYDRVGQDIISPLVSVKDLRELGVTLHLQLHSDRDSIPEVPAVYFCSPSDENLDRICQDLDNGIYDQYHLNFISPITRHKLEDLAASAIQSNSAVNIHKVFDQYLNFICLEDDLFIMKHQQSDSLSYYAINKADTKDTEMEVIMDNIVESLFSVFVTMGNVPFIRSTKGNAAEMVAKKLDKKLRENLWDARNNLFHGNTGQGGAFSFTRPMLILLDRNIDMATPLHHTWTYQALAHDVLDLSLNRAVVPENTNATVPIQKSNTRICDLDSKDPLWSEHKGSPFPTVAEAIQEDLDKYRSSEAEVKKLKSSMGLDAESDLALSMVSDNTQRLTNAVNSLPQLMEKKRLIDMHTTIATAILNAIKSRRLDSFFELEEKIMSKSTAVETKAVMDLITDPSAGTPEDKMRLFIIYYLCTSQFTDDEYEKFEGALVAANCDVKALSYMKRWKGFTKMSNQYEGGGTKTVSMFSKLVSQGSSFVMEGVKNLVVKKHKLPVSRIVESALSGSESAAELTWLDPRAARTDSAGRARSARQQPPTDAVVFLVGGGNYIEYHNLIDFAKQQAANGIPRKLIYGATTLPNGSQFLKQLSLLGEEIQ
Summary
Description
Non-vital for development.
Similarity
Belongs to the STXBP/unc-18/SEC1 family.
Keywords
Complete proteome
Cytoplasm
Membrane
Reference proteome
Repeat
Feature
chain Protein sly1 homolog
Uniprot
H9JLI0
A0A2A4ISF0
A0A2H1W8W1
A0A194PIA6
I4DKD1
A0A1E1W415
+ More
U5EM36 Q17JZ4 A0A1L8DVV5 A0A1Q3FAU3 D2A3F4 A0A1S4JCF2 A0A1W4X342 A0A2J7RNS8 A0A182KEL0 A0A1B6F8M0 A0A2M4ADP4 A0A2M4AD73 A0A2M3Z7R6 W5JW61 A0A2M4BGZ0 A0A182LH66 A0A182UTT0 A0A182HY28 A0A182WXF1 A0A182J7W9 B0WCI3 A0A182RVF9 A0A084VI10 U4UFV5 J3JWG1 A0A067RK32 A0A1B6CZ65 A0A2M4ADF9 A0A0A9Y8F8 A0A1J1IM12 N6TG38 A0A182LSU9 A0A182W1P6 A0A182FPL6 A0A224X933 A0A182N3A2 A0A182Y726 A0A1S4H6F8 A0A182P387 A0A0M4EE07 S4PAF6 T1PGS9 A0A0L0C8X1 A0A182R0D4 E2A440 A0A1W4UTS6 B5DJJ9 B4GSL4 A0A182U6U3 A0A1Y1NFQ2 A0A0P4VUA1 R4G4W4 B4KGU1 A0A336M193 B4LUE2 A0A3B0KA39 Q7QHU5 A0A1I8QD02 A0A0J9QUC5 B4NWQ5 B3NA17 Q24179 B4JAU2 B4I2L1 A0A336N046 E2BZI7 A0A0L7R233 A0A0J7P451 A0A1B0GMV8 B4MU03 B3N1Z5 A0A026W7W4 A0A0M8ZYC8 A0A151J9D2 A0A195CK26 A0A154PMR4 A0A0K8UIH7 A0A034VXV2 A0A088AVB0 F4WL32 A0A195B640 A0A158NZL2 O18637 E9IKW7 A0A195FC45 A0A151WLX0 A0A0A1XLL5 A0A1A9Y4F2 A0A1B0BGY7 A0A2A3ERD1 A0A232EJH6 A0A1A9V9F5 A0A310S9T6 A0A1A9ZN21
U5EM36 Q17JZ4 A0A1L8DVV5 A0A1Q3FAU3 D2A3F4 A0A1S4JCF2 A0A1W4X342 A0A2J7RNS8 A0A182KEL0 A0A1B6F8M0 A0A2M4ADP4 A0A2M4AD73 A0A2M3Z7R6 W5JW61 A0A2M4BGZ0 A0A182LH66 A0A182UTT0 A0A182HY28 A0A182WXF1 A0A182J7W9 B0WCI3 A0A182RVF9 A0A084VI10 U4UFV5 J3JWG1 A0A067RK32 A0A1B6CZ65 A0A2M4ADF9 A0A0A9Y8F8 A0A1J1IM12 N6TG38 A0A182LSU9 A0A182W1P6 A0A182FPL6 A0A224X933 A0A182N3A2 A0A182Y726 A0A1S4H6F8 A0A182P387 A0A0M4EE07 S4PAF6 T1PGS9 A0A0L0C8X1 A0A182R0D4 E2A440 A0A1W4UTS6 B5DJJ9 B4GSL4 A0A182U6U3 A0A1Y1NFQ2 A0A0P4VUA1 R4G4W4 B4KGU1 A0A336M193 B4LUE2 A0A3B0KA39 Q7QHU5 A0A1I8QD02 A0A0J9QUC5 B4NWQ5 B3NA17 Q24179 B4JAU2 B4I2L1 A0A336N046 E2BZI7 A0A0L7R233 A0A0J7P451 A0A1B0GMV8 B4MU03 B3N1Z5 A0A026W7W4 A0A0M8ZYC8 A0A151J9D2 A0A195CK26 A0A154PMR4 A0A0K8UIH7 A0A034VXV2 A0A088AVB0 F4WL32 A0A195B640 A0A158NZL2 O18637 E9IKW7 A0A195FC45 A0A151WLX0 A0A0A1XLL5 A0A1A9Y4F2 A0A1B0BGY7 A0A2A3ERD1 A0A232EJH6 A0A1A9V9F5 A0A310S9T6 A0A1A9ZN21
Pubmed
19121390
26354079
22651552
17510324
18362917
19820115
+ More
20920257 23761445 20966253 24438588 23537049 22516182 24845553 25401762 26823975 25244985 12364791 23622113 25315136 26108605 20798317 15632085 17994087 28004739 27129103 22936249 17550304 8675012 10731132 12537572 12537569 24508170 30249741 25348373 21719571 21347285 21282665 25830018 28648823
20920257 23761445 20966253 24438588 23537049 22516182 24845553 25401762 26823975 25244985 12364791 23622113 25315136 26108605 20798317 15632085 17994087 28004739 27129103 22936249 17550304 8675012 10731132 12537572 12537569 24508170 30249741 25348373 21719571 21347285 21282665 25830018 28648823
EMBL
BABH01005710
BABH01005711
BABH01005712
NWSH01008358
PCG62589.1
ODYU01006772
+ More
SOQ48924.1 KQ459603 KPI92778.1 AK401749 BAM18371.1 GDQN01009350 JAT81704.1 GANO01001166 JAB58705.1 CH477229 EAT47006.1 GFDF01003513 JAV10571.1 GFDL01010378 JAV24667.1 KQ971338 EFA01920.1 NEVH01002148 PNF42479.1 GECZ01023506 JAS46263.1 GGFK01005427 MBW38748.1 GGFK01005406 MBW38727.1 GGFM01003727 MBW24478.1 ADMH02000117 ETN67778.1 GGFJ01003184 MBW52325.1 APCN01004485 DS231888 EDS43554.1 ATLV01013254 KE524847 KFB37604.1 KB632278 ERL91253.1 BT127579 AEE62541.1 KK852424 KDR24206.1 GEDC01018538 JAS18760.1 GGFK01005492 MBW38813.1 GBHO01016221 GBRD01017639 GDHC01013782 JAG27383.1 JAG48188.1 JAQ04847.1 CVRI01000055 CRL01200.1 APGK01029087 KB740724 ENN79359.1 AXCM01007652 GFTR01007559 JAW08867.1 AAAB01008816 CP012523 ALC38230.1 GAIX01003284 JAA89276.1 KA647088 AFP61717.1 JRES01000753 KNC28726.1 AXCN02001120 GL436526 EFN71806.1 CH379061 EDY70471.1 CH479189 EDW25373.1 GEZM01009575 JAV94507.1 GDKW01000468 JAI56127.1 ACPB03020296 GAHY01000406 JAA77104.1 CH933807 EDW11141.1 UFQT01000238 SSX22217.1 CH940649 EDW64128.2 OUUW01000006 SPP81891.1 EAA05177.4 CM002910 KMY87683.1 CM000157 EDW87397.1 CH954177 EDV57480.1 U63852 AE014134 AY070995 BT022422 AAL48617.1 CH916368 EDW03900.1 CH480820 EDW54006.1 UFQS01002758 UFQT01002758 SSX14620.1 SSX34017.1 GL451627 EFN78885.1 KQ414666 KOC64935.1 LBMM01000153 KMR04714.1 AJVK01004227 AJVK01004228 AJVK01004229 CH963852 EDW75592.2 CH902667 EDV30605.1 KK107353 QOIP01000012 EZA52088.1 RLU16294.1 KQ435794 KOX73610.1 KQ979471 KYN21427.1 KQ977720 KYN00424.1 KQ434954 KZC12588.1 GDHF01025971 JAI26343.1 GAKP01011708 JAC47244.1 GL888207 EGI64995.1 KQ976587 KYM79725.1 ADTU01004724 U95037 GL764026 EFZ18853.1 KQ981685 KYN37983.1 KQ982944 KYQ48846.1 GBXI01014055 GBXI01002475 GBXI01002412 JAD00237.1 JAD11817.1 JAD11880.1 JXJN01014089 KZ288192 PBC34353.1 NNAY01004039 OXU18461.1 KQ766603 OAD53638.1
SOQ48924.1 KQ459603 KPI92778.1 AK401749 BAM18371.1 GDQN01009350 JAT81704.1 GANO01001166 JAB58705.1 CH477229 EAT47006.1 GFDF01003513 JAV10571.1 GFDL01010378 JAV24667.1 KQ971338 EFA01920.1 NEVH01002148 PNF42479.1 GECZ01023506 JAS46263.1 GGFK01005427 MBW38748.1 GGFK01005406 MBW38727.1 GGFM01003727 MBW24478.1 ADMH02000117 ETN67778.1 GGFJ01003184 MBW52325.1 APCN01004485 DS231888 EDS43554.1 ATLV01013254 KE524847 KFB37604.1 KB632278 ERL91253.1 BT127579 AEE62541.1 KK852424 KDR24206.1 GEDC01018538 JAS18760.1 GGFK01005492 MBW38813.1 GBHO01016221 GBRD01017639 GDHC01013782 JAG27383.1 JAG48188.1 JAQ04847.1 CVRI01000055 CRL01200.1 APGK01029087 KB740724 ENN79359.1 AXCM01007652 GFTR01007559 JAW08867.1 AAAB01008816 CP012523 ALC38230.1 GAIX01003284 JAA89276.1 KA647088 AFP61717.1 JRES01000753 KNC28726.1 AXCN02001120 GL436526 EFN71806.1 CH379061 EDY70471.1 CH479189 EDW25373.1 GEZM01009575 JAV94507.1 GDKW01000468 JAI56127.1 ACPB03020296 GAHY01000406 JAA77104.1 CH933807 EDW11141.1 UFQT01000238 SSX22217.1 CH940649 EDW64128.2 OUUW01000006 SPP81891.1 EAA05177.4 CM002910 KMY87683.1 CM000157 EDW87397.1 CH954177 EDV57480.1 U63852 AE014134 AY070995 BT022422 AAL48617.1 CH916368 EDW03900.1 CH480820 EDW54006.1 UFQS01002758 UFQT01002758 SSX14620.1 SSX34017.1 GL451627 EFN78885.1 KQ414666 KOC64935.1 LBMM01000153 KMR04714.1 AJVK01004227 AJVK01004228 AJVK01004229 CH963852 EDW75592.2 CH902667 EDV30605.1 KK107353 QOIP01000012 EZA52088.1 RLU16294.1 KQ435794 KOX73610.1 KQ979471 KYN21427.1 KQ977720 KYN00424.1 KQ434954 KZC12588.1 GDHF01025971 JAI26343.1 GAKP01011708 JAC47244.1 GL888207 EGI64995.1 KQ976587 KYM79725.1 ADTU01004724 U95037 GL764026 EFZ18853.1 KQ981685 KYN37983.1 KQ982944 KYQ48846.1 GBXI01014055 GBXI01002475 GBXI01002412 JAD00237.1 JAD11817.1 JAD11880.1 JXJN01014089 KZ288192 PBC34353.1 NNAY01004039 OXU18461.1 KQ766603 OAD53638.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000008820
UP000007266
UP000192223
+ More
UP000235965 UP000075881 UP000000673 UP000075882 UP000075903 UP000075840 UP000076407 UP000075880 UP000002320 UP000075900 UP000030765 UP000030742 UP000027135 UP000183832 UP000019118 UP000075883 UP000075920 UP000069272 UP000075884 UP000076408 UP000075885 UP000092553 UP000095301 UP000037069 UP000075886 UP000000311 UP000192221 UP000001819 UP000008744 UP000075902 UP000015103 UP000009192 UP000008792 UP000268350 UP000007062 UP000095300 UP000002282 UP000008711 UP000000803 UP000001070 UP000001292 UP000008237 UP000053825 UP000036403 UP000092462 UP000007798 UP000007801 UP000053097 UP000279307 UP000053105 UP000078492 UP000078542 UP000076502 UP000005203 UP000007755 UP000078540 UP000005205 UP000078541 UP000075809 UP000092443 UP000092460 UP000242457 UP000215335 UP000078200 UP000092445
UP000235965 UP000075881 UP000000673 UP000075882 UP000075903 UP000075840 UP000076407 UP000075880 UP000002320 UP000075900 UP000030765 UP000030742 UP000027135 UP000183832 UP000019118 UP000075883 UP000075920 UP000069272 UP000075884 UP000076408 UP000075885 UP000092553 UP000095301 UP000037069 UP000075886 UP000000311 UP000192221 UP000001819 UP000008744 UP000075902 UP000015103 UP000009192 UP000008792 UP000268350 UP000007062 UP000095300 UP000002282 UP000008711 UP000000803 UP000001070 UP000001292 UP000008237 UP000053825 UP000036403 UP000092462 UP000007798 UP000007801 UP000053097 UP000279307 UP000053105 UP000078492 UP000078542 UP000076502 UP000005203 UP000007755 UP000078540 UP000005205 UP000078541 UP000075809 UP000092443 UP000092460 UP000242457 UP000215335 UP000078200 UP000092445
Interpro
IPR001619
Sec1-like
+ More
IPR036045 Sec1-like_sf
IPR027482 Sec-1-like_dom2
IPR000412 ABC_2_transport
IPR027417 P-loop_NTPase
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR003593 AAA+_ATPase
IPR000698 Arrestin
IPR011022 Arrestin_C-like
IPR014756 Ig_E-set
IPR014752 Arrestin_C
IPR014753 Arrestin_N
IPR011021 Arrestin-like_N
IPR036045 Sec1-like_sf
IPR027482 Sec-1-like_dom2
IPR000412 ABC_2_transport
IPR027417 P-loop_NTPase
IPR003439 ABC_transporter-like
IPR017871 ABC_transporter_CS
IPR003593 AAA+_ATPase
IPR000698 Arrestin
IPR011022 Arrestin_C-like
IPR014756 Ig_E-set
IPR014752 Arrestin_C
IPR014753 Arrestin_N
IPR011021 Arrestin-like_N
Gene 3D
ProteinModelPortal
H9JLI0
A0A2A4ISF0
A0A2H1W8W1
A0A194PIA6
I4DKD1
A0A1E1W415
+ More
U5EM36 Q17JZ4 A0A1L8DVV5 A0A1Q3FAU3 D2A3F4 A0A1S4JCF2 A0A1W4X342 A0A2J7RNS8 A0A182KEL0 A0A1B6F8M0 A0A2M4ADP4 A0A2M4AD73 A0A2M3Z7R6 W5JW61 A0A2M4BGZ0 A0A182LH66 A0A182UTT0 A0A182HY28 A0A182WXF1 A0A182J7W9 B0WCI3 A0A182RVF9 A0A084VI10 U4UFV5 J3JWG1 A0A067RK32 A0A1B6CZ65 A0A2M4ADF9 A0A0A9Y8F8 A0A1J1IM12 N6TG38 A0A182LSU9 A0A182W1P6 A0A182FPL6 A0A224X933 A0A182N3A2 A0A182Y726 A0A1S4H6F8 A0A182P387 A0A0M4EE07 S4PAF6 T1PGS9 A0A0L0C8X1 A0A182R0D4 E2A440 A0A1W4UTS6 B5DJJ9 B4GSL4 A0A182U6U3 A0A1Y1NFQ2 A0A0P4VUA1 R4G4W4 B4KGU1 A0A336M193 B4LUE2 A0A3B0KA39 Q7QHU5 A0A1I8QD02 A0A0J9QUC5 B4NWQ5 B3NA17 Q24179 B4JAU2 B4I2L1 A0A336N046 E2BZI7 A0A0L7R233 A0A0J7P451 A0A1B0GMV8 B4MU03 B3N1Z5 A0A026W7W4 A0A0M8ZYC8 A0A151J9D2 A0A195CK26 A0A154PMR4 A0A0K8UIH7 A0A034VXV2 A0A088AVB0 F4WL32 A0A195B640 A0A158NZL2 O18637 E9IKW7 A0A195FC45 A0A151WLX0 A0A0A1XLL5 A0A1A9Y4F2 A0A1B0BGY7 A0A2A3ERD1 A0A232EJH6 A0A1A9V9F5 A0A310S9T6 A0A1A9ZN21
U5EM36 Q17JZ4 A0A1L8DVV5 A0A1Q3FAU3 D2A3F4 A0A1S4JCF2 A0A1W4X342 A0A2J7RNS8 A0A182KEL0 A0A1B6F8M0 A0A2M4ADP4 A0A2M4AD73 A0A2M3Z7R6 W5JW61 A0A2M4BGZ0 A0A182LH66 A0A182UTT0 A0A182HY28 A0A182WXF1 A0A182J7W9 B0WCI3 A0A182RVF9 A0A084VI10 U4UFV5 J3JWG1 A0A067RK32 A0A1B6CZ65 A0A2M4ADF9 A0A0A9Y8F8 A0A1J1IM12 N6TG38 A0A182LSU9 A0A182W1P6 A0A182FPL6 A0A224X933 A0A182N3A2 A0A182Y726 A0A1S4H6F8 A0A182P387 A0A0M4EE07 S4PAF6 T1PGS9 A0A0L0C8X1 A0A182R0D4 E2A440 A0A1W4UTS6 B5DJJ9 B4GSL4 A0A182U6U3 A0A1Y1NFQ2 A0A0P4VUA1 R4G4W4 B4KGU1 A0A336M193 B4LUE2 A0A3B0KA39 Q7QHU5 A0A1I8QD02 A0A0J9QUC5 B4NWQ5 B3NA17 Q24179 B4JAU2 B4I2L1 A0A336N046 E2BZI7 A0A0L7R233 A0A0J7P451 A0A1B0GMV8 B4MU03 B3N1Z5 A0A026W7W4 A0A0M8ZYC8 A0A151J9D2 A0A195CK26 A0A154PMR4 A0A0K8UIH7 A0A034VXV2 A0A088AVB0 F4WL32 A0A195B640 A0A158NZL2 O18637 E9IKW7 A0A195FC45 A0A151WLX0 A0A0A1XLL5 A0A1A9Y4F2 A0A1B0BGY7 A0A2A3ERD1 A0A232EJH6 A0A1A9V9F5 A0A310S9T6 A0A1A9ZN21
PDB
1MQS
E-value=4.30649e-84,
Score=795
Ontologies
GO
GO:0006904
GO:0016021
GO:0016887
GO:0055085
GO:0043190
GO:0005524
GO:0007165
GO:0000902
GO:0048190
GO:0006892
GO:0019905
GO:0009636
GO:0001666
GO:0000149
GO:0060628
GO:0005801
GO:0016192
GO:0044877
GO:0005798
GO:0006890
GO:0005886
GO:0005737
GO:0016020
GO:0006468
GO:0019843
GO:0005622
GO:0004656
GO:0005783
GO:0016702
GO:0016295
GO:0016296
PANTHER
Topology
Subcellular location
Cytoplasm
Membrane
Membrane
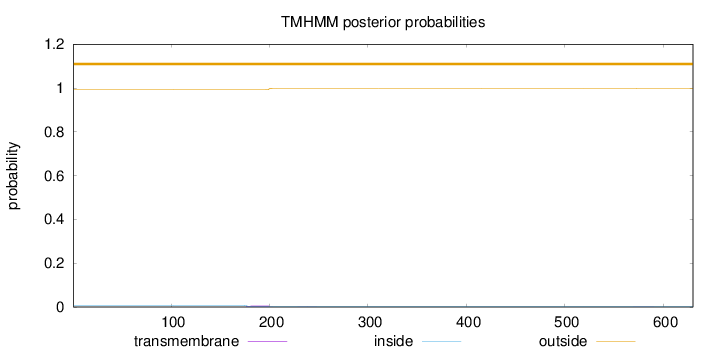
Length:
630
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15827
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00769
outside
1 - 630
Population Genetic Test Statistics
Pi
143.997218
Theta
155.259654
Tajima's D
-0.407398
CLR
1.880139
CSRT
0.267336633168342
Interpretation
Uncertain
