Gene
KWMTBOMO07204 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010572
Annotation
calcium/calmodulin-dependent_protein_kinase_type_1_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.242 Nuclear Reliability : 2.152
Sequence
CDS
ATGCCTCTTTTCGGTAAAAAAGATTTCGGAAGGAAATCTAAAAAAGATGGAAAAGACTCTGAAAAACAACCGTCTATTGAAGACAAATACATTGTAAAGGACCTTTTGGGTACGGGGGCCTTTTCTGAGGTGCGTTTGATAGAGAGCAAAGAAAGCGGCCAGCTGTTTGCTTGCAAAATCATTGATAAAAAAGCCCTTAAAGGTAAAGAAGATTCCCTTGAAAATGAAATTAGGGTATTAAAGCGATTTAGTGAACAAGGTAAGGGTGAAGGTGGGGAAAAGAAAGTTTTTTCTCATCCGAATATAGTACAGCTACTCGAAACATACGAGGACAAAAATAAAGTTTACCTTGTCATGGAGCTAGTAACAGGTGGTGAATTATTTGATAGAATTGTTGAAAAGGGTTCCTATACTGAAAAAGATGCGTCCAATTTGATTAGACAGGTGTTGGAAGCTGTTGATTATATGCATTCTCAGGGAGTTGTGCACAGAGATTTGAAGCCTGAAAATCTTCTTTACTACAGCACTGAGGAAGACAGTAAGATAATGATTAGTGATTTTGGGTTATCTAAGATTGAGGATTCTGGCATAATGGCCACAGCATGTGGTACACCAGGCTATGTAGCACCTGAAGTATTGGCACAGAAGCCATATGGTAAGGCTGTTGATGTCTGGAGTATAGGAGTCATATCATACATTTTGCTGTGTGGATATCCTCCATTTTATGACGAGAATGATGCAAACTTATTTGCCCAGATTCTAAAAGGCGATTTTGAATTTGATTCTCCTTATTGGGATGATATAAGCGATTCAGCTAAAGATTTTATCCGACATCTTATGTGTGTTGATGTAGAAAAGAGATACACTTGCAAGCAAGCCTTGGCTCATCCATGGATTTCTGGTAATGCTGCTAGCAATAAGAACATTCACGGTACAGTTTCGGAACAACTGAAGAAGAACTTTGCAAAGTCCCGCTGGAAACAAGCCTACCATGCTACAACAGTCATTAGACAGATGCAGAAGATGATTTTGAACAGCAGTGGTTCAAGTCGTTCTGCCAGTCAATCTCAGCCAAAACAATAG
Protein
MPLFGKKDFGRKSKKDGKDSEKQPSIEDKYIVKDLLGTGAFSEVRLIESKESGQLFACKIIDKKALKGKEDSLENEIRVLKRFSEQGKGEGGEKKVFSHPNIVQLLETYEDKNKVYLVMELVTGGELFDRIVEKGSYTEKDASNLIRQVLEAVDYMHSQGVVHRDLKPENLLYYSTEEDSKIMISDFGLSKIEDSGIMATACGTPGYVAPEVLAQKPYGKAVDVWSIGVISYILLCGYPPFYDENDANLFAQILKGDFEFDSPYWDDISDSAKDFIRHLMCVDVEKRYTCKQALAHPWISGNAASNKNIHGTVSEQLKKNFAKSRWKQAYHATTVIRQMQKMILNSSGSSRSASQSQPKQ
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
H9JM20
A0A194R057
A0A194PHI5
A0A3S2M9V8
A0A212FL37
A0A2H1W746
+ More
A0A2A4ITY0 A0A0L7LFZ0 Q16Q29 A0A1S4FTK5 T1DED3 A0A1Q3G4I5 A0A182GGI9 A0A182KE01 A0A182NN66 A0A182YNZ6 A0A182Q3J5 A0A084WJI8 A0A182WS06 A0A182U1Y1 A0A182VMQ4 A0A182ICW4 A0A182JI42 A0A182MAL8 Q7QI42 A0A2M4AAL3 A0A182FCY6 A0A2M4BPF5 A0A2M3ZGI2 A0A2M3Z1M2 W5JHN5 U5EY85 A0A182RLG6 A0A1J1IDF2 A0A195EUE5 A0A1L8DL44 A0A0T6AZ15 A0A336JXH2 A0A1L8DKF4 A0A0M3QYS8 A0A0K8TMZ5 Q29CP9 B4H883 K7J7B9 Q7JMV3 E2ADT1 B4JZV5 B4PW07 B4L7C2 B3P9T4 A0A195B0Y3 A0A1I8MRJ3 A0A1I8PV45 B4NHM4 F4WPE0 A4V110 E2BN34 A0A1S6J0Z6 A0A0R3NVM0 A0A3B0K4D0 A0A0Q5STB0 A0A1B6EE84 A0A1Y1N2A1 A0A0R1E6R3 E0W2P0 A0A026WPW6 B3MY55 A0A158NQZ1 A0A0Q9WVJ4 A0A0Q9XFG4 A0A088A4Q2 A0A1B6LT40 B4MF09 A0A154PNC1 A0A0P8XZL1 D6W6N7 A0A1B6L934 A0A1A9X532 A0A0Q9WKZ2 A0A0A1X7V0 A0A1A9VYH4 A0A1A9Z3J0 Q2PQP0 D0Z7B0 A0A0N8E1P4 A0A1B0B9E4 A0A034WX84 A0A0P5VN27 A0A0P5W7D2 A0A2S2QQ24 A0A0K8U843 A0A131YU38 A0A1W4VH76 L7M0B8 A0A0P6F5N4 A0A2A3ED62 A0A0A1XM67 E9HCA1 A0A1B6C8G6 A0A1W4VHU1
A0A2A4ITY0 A0A0L7LFZ0 Q16Q29 A0A1S4FTK5 T1DED3 A0A1Q3G4I5 A0A182GGI9 A0A182KE01 A0A182NN66 A0A182YNZ6 A0A182Q3J5 A0A084WJI8 A0A182WS06 A0A182U1Y1 A0A182VMQ4 A0A182ICW4 A0A182JI42 A0A182MAL8 Q7QI42 A0A2M4AAL3 A0A182FCY6 A0A2M4BPF5 A0A2M3ZGI2 A0A2M3Z1M2 W5JHN5 U5EY85 A0A182RLG6 A0A1J1IDF2 A0A195EUE5 A0A1L8DL44 A0A0T6AZ15 A0A336JXH2 A0A1L8DKF4 A0A0M3QYS8 A0A0K8TMZ5 Q29CP9 B4H883 K7J7B9 Q7JMV3 E2ADT1 B4JZV5 B4PW07 B4L7C2 B3P9T4 A0A195B0Y3 A0A1I8MRJ3 A0A1I8PV45 B4NHM4 F4WPE0 A4V110 E2BN34 A0A1S6J0Z6 A0A0R3NVM0 A0A3B0K4D0 A0A0Q5STB0 A0A1B6EE84 A0A1Y1N2A1 A0A0R1E6R3 E0W2P0 A0A026WPW6 B3MY55 A0A158NQZ1 A0A0Q9WVJ4 A0A0Q9XFG4 A0A088A4Q2 A0A1B6LT40 B4MF09 A0A154PNC1 A0A0P8XZL1 D6W6N7 A0A1B6L934 A0A1A9X532 A0A0Q9WKZ2 A0A0A1X7V0 A0A1A9VYH4 A0A1A9Z3J0 Q2PQP0 D0Z7B0 A0A0N8E1P4 A0A1B0B9E4 A0A034WX84 A0A0P5VN27 A0A0P5W7D2 A0A2S2QQ24 A0A0K8U843 A0A131YU38 A0A1W4VH76 L7M0B8 A0A0P6F5N4 A0A2A3ED62 A0A0A1XM67 E9HCA1 A0A1B6C8G6 A0A1W4VHU1
Pubmed
19121390
26354079
22118469
26227816
17510324
24330624
+ More
26483478 25244985 24438588 12364791 14747013 17210077 20920257 23761445 26369729 15632085 17994087 20075255 9813038 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17550304 25315136 18057021 21719571 27414796 28004739 20566863 24508170 30249741 21347285 18362917 19820115 25830018 16907828 20353571 25348373 26830274 25576852 21292972
26483478 25244985 24438588 12364791 14747013 17210077 20920257 23761445 26369729 15632085 17994087 20075255 9813038 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 17550304 25315136 18057021 21719571 27414796 28004739 20566863 24508170 30249741 21347285 18362917 19820115 25830018 16907828 20353571 25348373 26830274 25576852 21292972
EMBL
BABH01005710
KQ460883
KPJ11183.1
KQ459603
KPI92777.1
RSAL01000003
+ More
RVE54757.1 AGBW02007871 OWR54463.1 ODYU01006772 SOQ48925.1 NWSH01008358 PCG62590.1 JTDY01001258 KOB74377.1 CH477765 EAT36480.1 GALA01001077 JAA93775.1 GFDL01000341 JAV34704.1 JXUM01061825 KQ562170 KXJ76517.1 AXCN02001950 ATLV01024035 KE525348 KFB50382.1 APCN01006051 AXCM01006346 AAAB01008810 EAA04816.4 GGFK01004459 MBW37780.1 GGFJ01005825 MBW54966.1 GGFM01006878 MBW27629.1 GGFM01001640 MBW22391.1 ADMH02001370 ETN62818.1 GANO01000523 JAB59348.1 CVRI01000047 CRK97588.1 KQ981965 KYN31875.1 GFDF01007010 JAV07074.1 LJIG01022477 KRT80364.1 UFQS01000005 UFQT01000005 SSW96901.1 SSX17288.1 GFDF01007131 JAV06953.1 CP012527 ALC48139.1 GDAI01001866 JAI15737.1 CH475436 EAL29266.2 CH479221 EDW34883.1 AE014135 BT001637 Y17917 AAF59343.1 AAF59344.2 AAN06531.1 AAN71392.1 CAA76937.1 GL438820 EFN68443.1 CH916380 EDV90971.1 CM000161 EDW99312.1 CH933813 EDW10916.1 CH954184 EDV45247.1 KQ976691 KYM77952.1 CH964272 EDW83593.1 KRF99829.1 GL888243 EGI64035.1 AAN06532.2 GL449382 EFN82938.1 KU764449 AQS60695.1 KRT05154.1 OUUW01000017 SPP89005.1 KQS21499.1 GEDC01001114 JAS36184.1 GEZM01019349 JAV89697.1 KRK04872.1 AAZO01007265 DS235879 EEB19896.1 KK107139 QOIP01000003 EZA57711.1 RLU24342.1 CH902631 EDV34087.1 ADTU01023707 ADTU01023708 KRF99828.1 KRG07275.1 GEBQ01013188 JAT26789.1 CH940665 EDW71110.1 KRF85364.1 KRF85365.1 KQ434999 KZC13366.1 KPU74971.1 KQ971307 EFA10976.1 GEBQ01019755 JAT20222.1 KRF85363.1 GBXI01007539 JAD06753.1 CCAG010016063 DQ307183 EZ423367 ABC25083.1 ADD19643.1 CM000831 ACY70523.1 GDIQ01070516 JAN24221.1 JXJN01010368 GAKP01000544 JAC58408.1 GDIP01097806 JAM05909.1 GDIP01090156 JAM13559.1 GGMS01010427 MBY79630.1 GDHF01029571 JAI22743.1 GEDV01006080 JAP82477.1 GACK01007722 JAA57312.1 GDIQ01054218 JAN40519.1 KZ288295 PBC28961.1 GBXI01002337 JAD11955.1 GL732619 EFX70647.1 GEDC01027768 JAS09530.1
RVE54757.1 AGBW02007871 OWR54463.1 ODYU01006772 SOQ48925.1 NWSH01008358 PCG62590.1 JTDY01001258 KOB74377.1 CH477765 EAT36480.1 GALA01001077 JAA93775.1 GFDL01000341 JAV34704.1 JXUM01061825 KQ562170 KXJ76517.1 AXCN02001950 ATLV01024035 KE525348 KFB50382.1 APCN01006051 AXCM01006346 AAAB01008810 EAA04816.4 GGFK01004459 MBW37780.1 GGFJ01005825 MBW54966.1 GGFM01006878 MBW27629.1 GGFM01001640 MBW22391.1 ADMH02001370 ETN62818.1 GANO01000523 JAB59348.1 CVRI01000047 CRK97588.1 KQ981965 KYN31875.1 GFDF01007010 JAV07074.1 LJIG01022477 KRT80364.1 UFQS01000005 UFQT01000005 SSW96901.1 SSX17288.1 GFDF01007131 JAV06953.1 CP012527 ALC48139.1 GDAI01001866 JAI15737.1 CH475436 EAL29266.2 CH479221 EDW34883.1 AE014135 BT001637 Y17917 AAF59343.1 AAF59344.2 AAN06531.1 AAN71392.1 CAA76937.1 GL438820 EFN68443.1 CH916380 EDV90971.1 CM000161 EDW99312.1 CH933813 EDW10916.1 CH954184 EDV45247.1 KQ976691 KYM77952.1 CH964272 EDW83593.1 KRF99829.1 GL888243 EGI64035.1 AAN06532.2 GL449382 EFN82938.1 KU764449 AQS60695.1 KRT05154.1 OUUW01000017 SPP89005.1 KQS21499.1 GEDC01001114 JAS36184.1 GEZM01019349 JAV89697.1 KRK04872.1 AAZO01007265 DS235879 EEB19896.1 KK107139 QOIP01000003 EZA57711.1 RLU24342.1 CH902631 EDV34087.1 ADTU01023707 ADTU01023708 KRF99828.1 KRG07275.1 GEBQ01013188 JAT26789.1 CH940665 EDW71110.1 KRF85364.1 KRF85365.1 KQ434999 KZC13366.1 KPU74971.1 KQ971307 EFA10976.1 GEBQ01019755 JAT20222.1 KRF85363.1 GBXI01007539 JAD06753.1 CCAG010016063 DQ307183 EZ423367 ABC25083.1 ADD19643.1 CM000831 ACY70523.1 GDIQ01070516 JAN24221.1 JXJN01010368 GAKP01000544 JAC58408.1 GDIP01097806 JAM05909.1 GDIP01090156 JAM13559.1 GGMS01010427 MBY79630.1 GDHF01029571 JAI22743.1 GEDV01006080 JAP82477.1 GACK01007722 JAA57312.1 GDIQ01054218 JAN40519.1 KZ288295 PBC28961.1 GBXI01002337 JAD11955.1 GL732619 EFX70647.1 GEDC01027768 JAS09530.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
+ More
UP000037510 UP000008820 UP000069940 UP000249989 UP000075881 UP000075884 UP000076408 UP000075886 UP000030765 UP000076407 UP000075902 UP000075903 UP000075840 UP000075880 UP000075883 UP000007062 UP000069272 UP000000673 UP000075900 UP000183832 UP000078541 UP000092553 UP000001819 UP000008744 UP000002358 UP000000803 UP000000311 UP000001070 UP000002282 UP000009192 UP000008711 UP000078540 UP000095301 UP000095300 UP000007798 UP000007755 UP000008237 UP000268350 UP000009046 UP000053097 UP000279307 UP000007801 UP000005205 UP000005203 UP000008792 UP000076502 UP000007266 UP000091820 UP000078200 UP000092445 UP000092444 UP000092460 UP000192221 UP000242457 UP000000305
UP000037510 UP000008820 UP000069940 UP000249989 UP000075881 UP000075884 UP000076408 UP000075886 UP000030765 UP000076407 UP000075902 UP000075903 UP000075840 UP000075880 UP000075883 UP000007062 UP000069272 UP000000673 UP000075900 UP000183832 UP000078541 UP000092553 UP000001819 UP000008744 UP000002358 UP000000803 UP000000311 UP000001070 UP000002282 UP000009192 UP000008711 UP000078540 UP000095301 UP000095300 UP000007798 UP000007755 UP000008237 UP000268350 UP000009046 UP000053097 UP000279307 UP000007801 UP000005205 UP000005203 UP000008792 UP000076502 UP000007266 UP000091820 UP000078200 UP000092445 UP000092444 UP000092460 UP000192221 UP000242457 UP000000305
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JM20
A0A194R057
A0A194PHI5
A0A3S2M9V8
A0A212FL37
A0A2H1W746
+ More
A0A2A4ITY0 A0A0L7LFZ0 Q16Q29 A0A1S4FTK5 T1DED3 A0A1Q3G4I5 A0A182GGI9 A0A182KE01 A0A182NN66 A0A182YNZ6 A0A182Q3J5 A0A084WJI8 A0A182WS06 A0A182U1Y1 A0A182VMQ4 A0A182ICW4 A0A182JI42 A0A182MAL8 Q7QI42 A0A2M4AAL3 A0A182FCY6 A0A2M4BPF5 A0A2M3ZGI2 A0A2M3Z1M2 W5JHN5 U5EY85 A0A182RLG6 A0A1J1IDF2 A0A195EUE5 A0A1L8DL44 A0A0T6AZ15 A0A336JXH2 A0A1L8DKF4 A0A0M3QYS8 A0A0K8TMZ5 Q29CP9 B4H883 K7J7B9 Q7JMV3 E2ADT1 B4JZV5 B4PW07 B4L7C2 B3P9T4 A0A195B0Y3 A0A1I8MRJ3 A0A1I8PV45 B4NHM4 F4WPE0 A4V110 E2BN34 A0A1S6J0Z6 A0A0R3NVM0 A0A3B0K4D0 A0A0Q5STB0 A0A1B6EE84 A0A1Y1N2A1 A0A0R1E6R3 E0W2P0 A0A026WPW6 B3MY55 A0A158NQZ1 A0A0Q9WVJ4 A0A0Q9XFG4 A0A088A4Q2 A0A1B6LT40 B4MF09 A0A154PNC1 A0A0P8XZL1 D6W6N7 A0A1B6L934 A0A1A9X532 A0A0Q9WKZ2 A0A0A1X7V0 A0A1A9VYH4 A0A1A9Z3J0 Q2PQP0 D0Z7B0 A0A0N8E1P4 A0A1B0B9E4 A0A034WX84 A0A0P5VN27 A0A0P5W7D2 A0A2S2QQ24 A0A0K8U843 A0A131YU38 A0A1W4VH76 L7M0B8 A0A0P6F5N4 A0A2A3ED62 A0A0A1XM67 E9HCA1 A0A1B6C8G6 A0A1W4VHU1
A0A2A4ITY0 A0A0L7LFZ0 Q16Q29 A0A1S4FTK5 T1DED3 A0A1Q3G4I5 A0A182GGI9 A0A182KE01 A0A182NN66 A0A182YNZ6 A0A182Q3J5 A0A084WJI8 A0A182WS06 A0A182U1Y1 A0A182VMQ4 A0A182ICW4 A0A182JI42 A0A182MAL8 Q7QI42 A0A2M4AAL3 A0A182FCY6 A0A2M4BPF5 A0A2M3ZGI2 A0A2M3Z1M2 W5JHN5 U5EY85 A0A182RLG6 A0A1J1IDF2 A0A195EUE5 A0A1L8DL44 A0A0T6AZ15 A0A336JXH2 A0A1L8DKF4 A0A0M3QYS8 A0A0K8TMZ5 Q29CP9 B4H883 K7J7B9 Q7JMV3 E2ADT1 B4JZV5 B4PW07 B4L7C2 B3P9T4 A0A195B0Y3 A0A1I8MRJ3 A0A1I8PV45 B4NHM4 F4WPE0 A4V110 E2BN34 A0A1S6J0Z6 A0A0R3NVM0 A0A3B0K4D0 A0A0Q5STB0 A0A1B6EE84 A0A1Y1N2A1 A0A0R1E6R3 E0W2P0 A0A026WPW6 B3MY55 A0A158NQZ1 A0A0Q9WVJ4 A0A0Q9XFG4 A0A088A4Q2 A0A1B6LT40 B4MF09 A0A154PNC1 A0A0P8XZL1 D6W6N7 A0A1B6L934 A0A1A9X532 A0A0Q9WKZ2 A0A0A1X7V0 A0A1A9VYH4 A0A1A9Z3J0 Q2PQP0 D0Z7B0 A0A0N8E1P4 A0A1B0B9E4 A0A034WX84 A0A0P5VN27 A0A0P5W7D2 A0A2S2QQ24 A0A0K8U843 A0A131YU38 A0A1W4VH76 L7M0B8 A0A0P6F5N4 A0A2A3ED62 A0A0A1XM67 E9HCA1 A0A1B6C8G6 A0A1W4VHU1
PDB
1A06
E-value=1.20677e-124,
Score=1142
Ontologies
GO
Topology
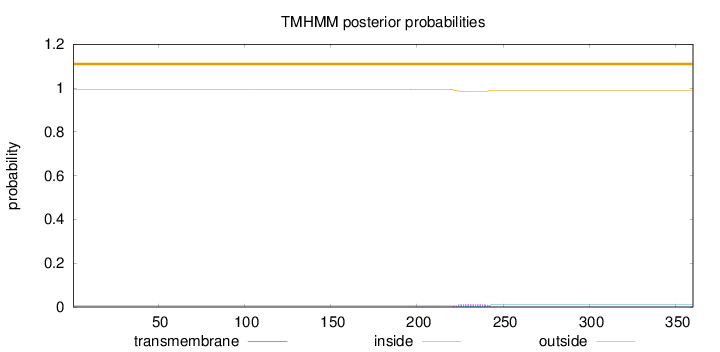
Length:
360
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23551
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00564
outside
1 - 360
Population Genetic Test Statistics
Pi
212.939574
Theta
213.152585
Tajima's D
-0.093041
CLR
0
CSRT
0.349082545872706
Interpretation
Uncertain
