Gene
KWMTBOMO07198
Pre Gene Modal
BGIBMGA010570
Annotation
PREDICTED:_T-cell_activation_inhibitor?_mitochondrial_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.487 Nuclear Reliability : 1.776
Sequence
CDS
ATGTATTGTAGACTAAATATAAGATGTATTTGCGCAAATGGAGTAACCAGCAGATTTCTCAGTACAGCAGAAATCTCTACAGCACTTCGACCATTTTACTTCAGTGTACATCCAGATCTTTTTGGAAAATATCCAGAACAAAGGAAAATAAACGAACAGTCACTGCAACAGCTGAGTGCTTTTTTAGAAGCTCAACAATCTAGAAGGAGAATGGTTGTACCGCCTTTGCCATTCTATTTGCGACAAAAAAACACAGCAGAAGGTGAATTTAAATTAGTACAAATCCAATTAAATGGCAGCGATACAAGGGACACAGTTGTTAAAGTACTGAACGCATGTAATATATCAACAAGCTATGTGGACAAGATACCTCAAGCTCCTCTAAAAGAAGATTCATTTAGCAAACCTGATTTCATGAAGGCATACCAACGCTATAGTACAGAATTTGAAAATGCAACTGAAATAAAAAAGAAAGTAGAAGAAAAGAAAGCAATCGACAATATTGTAGACTGGGTCTACGAGAACAGTGCTGTGGCCCGTGAAAAATATGAGCTGATGAGCGCAACAAGAGACCAAGTGAAGAGCCTGATTGATCACCTATGCAAAACTTACGGTATTAAAGAAGTGAAATATGATAATGGCTGGAATATCAGTCATATCAGAGGAGCTTTGCAGAGTCTAGTTTCACTTGCTACGCAACATTCCAATATTATGGATAATTTACAAGGCAGGACAATTGCACTGGGTCAATTCACCGGAGTGAGCTTGGATGGAGATGTATATCTTAACATTATAGATGTCAGAAATGAGTGGTTATCATTAATAAAGAAAGTAAGGCAAGAAGACAGCGCGCTGACTATGATACCTAGTTATGAGAAGGCTTTATCCAGTATCTTACGTGATATTCATATACGAAGAAGAAAATTCATGCCGAAGGTTTCGGCGGCGCTATATTGTAGTCATCTGAGACAGCTAATAACGTCCATAGGCGATTTTTACGGACGGGGAAACAAATTTCCTCCGTCTGTACCGGAATCTCTTAGCAAATACGAAATCGTCGTTGAACCGGAGGCAGGCCCTCTGATGGTGTCGCCGACTGGACAGTTCATCACTCCGTCGTCGTGTCCAGCTGACGAGCTAATAGCATACATAAATAATCATTTAGACGAAGCGACTTTACTTCTAACCGAATATAGCATAAATAAACACGTCGAGAAAGCATTACACAAAGAGGTAAAAGAAAGATTTGGACTACTAGAACTACACAAGGACGATAGTATAACTCCAGGACTAATGATTTCGTGTTGTCAACGTCTATTGACTAGAACGGATAAACTTAATGCGAATCTACGAGGTAACATATTGTACGTAACACATTATTACTCTGTGTTAACTGAAGGCATTTTATGTATACCGTGGAATTTTAAATAA
Protein
MYCRLNIRCICANGVTSRFLSTAEISTALRPFYFSVHPDLFGKYPEQRKINEQSLQQLSAFLEAQQSRRRMVVPPLPFYLRQKNTAEGEFKLVQIQLNGSDTRDTVVKVLNACNISTSYVDKIPQAPLKEDSFSKPDFMKAYQRYSTEFENATEIKKKVEEKKAIDNIVDWVYENSAVAREKYELMSATRDQVKSLIDHLCKTYGIKEVKYDNGWNISHIRGALQSLVSLATQHSNIMDNLQGRTIALGQFTGVSLDGDVYLNIIDVRNEWLSLIKKVRQEDSALTMIPSYEKALSSILRDIHIRRRKFMPKVSAALYCSHLRQLITSIGDFYGRGNKFPPSVPESLSKYEIVVEPEAGPLMVSPTGQFITPSSCPADELIAYINNHLDEATLLLTEYSINKHVEKALHKEVKERFGLLELHKDDSITPGLMISCCQRLLTRTDKLNANLRGNILYVTHYYSVLTEGILCIPWNFK
Summary
Uniprot
A0A194R5Q0
A0A212FL33
A0A2H1X2K8
A0A2A4JS76
A0A194PJ71
I4DJW2
+ More
K7J0J1 A0A232F3I7 A0A2J7RQ39 A0A154PQ94 A0A3L8DWZ2 E2AZ37 A0A067QYM4 A0A195FSN7 A0A087ZYU2 A0A2A3EG43 A0A151WR73 E2CAD5 A0A151J3T4 A0A026W4U8 A0A139WH37 A0A158NYY2 A0A195D0W2 A0A1W4WSN9 A0A151I4P8 A0A0C9R6R8 F4W567 A0A1B6M2B5 T1HXI5 A0A1Y1K6N3 A0A1B6EYD2 A0A1B6DS34 A0A182GEQ5 A0A1J1HEH2 Q16YL5 A0A1S4FJS7 A0A2M4BKB5 A0A2M4BKD3 A0A2M4BL53 A0A1L8E2U1 A0A182NF04 A0A1L8E344 Q5TND9 A0A182STQ0 A0A182YFB5 A0A0L7RH96 W5JKY5 A0A182WH27 A0A182V7F0 A0A1B0GQI2 A0A2M4AHN4 A0A2M4AHB0 A0A084WBY5 A0A336M257 A0A1Q3FC71 A0A1Y9HEF4 A0A182F5D6 A0A182Q5V3 A0A182JF51 A0A182KUL8 A0A182NZW3 A0A182TZY2 A0A0A9Z0P0 A0A1S4H067 A0A182WYP7 B0W1H6 A0A182M1Q6 E9IUX8 A0A1I8N1U1 T1PAE2 A0A182I524 A0A1D2NAU9 A0A0P4Z6L1 A0A0P5R1V6 A0A164ZZF4 A0A0P4ZKJ0 A0A0N7ZML8 A0A224XGU8 A0A0P5YDW3 A0A0P6A9Z7 A0A0N8AEL8 B4LDD9 A0A0P5TMZ2 A0A0N8CYD1 B4L9P7 R4G8X1 A0A146LPN9 A0A226EX63 A0A1A9Y2A5 A0A1B0C6B3 A0A2M3Z972 A0A0P6HW72 A0A0J7L9Y8 A0A1B0GCR2 A0A0P5LJ37 A0A0P5WEK5 A0A0L0CG71 B4IZY8 A0A0P6DXN4
K7J0J1 A0A232F3I7 A0A2J7RQ39 A0A154PQ94 A0A3L8DWZ2 E2AZ37 A0A067QYM4 A0A195FSN7 A0A087ZYU2 A0A2A3EG43 A0A151WR73 E2CAD5 A0A151J3T4 A0A026W4U8 A0A139WH37 A0A158NYY2 A0A195D0W2 A0A1W4WSN9 A0A151I4P8 A0A0C9R6R8 F4W567 A0A1B6M2B5 T1HXI5 A0A1Y1K6N3 A0A1B6EYD2 A0A1B6DS34 A0A182GEQ5 A0A1J1HEH2 Q16YL5 A0A1S4FJS7 A0A2M4BKB5 A0A2M4BKD3 A0A2M4BL53 A0A1L8E2U1 A0A182NF04 A0A1L8E344 Q5TND9 A0A182STQ0 A0A182YFB5 A0A0L7RH96 W5JKY5 A0A182WH27 A0A182V7F0 A0A1B0GQI2 A0A2M4AHN4 A0A2M4AHB0 A0A084WBY5 A0A336M257 A0A1Q3FC71 A0A1Y9HEF4 A0A182F5D6 A0A182Q5V3 A0A182JF51 A0A182KUL8 A0A182NZW3 A0A182TZY2 A0A0A9Z0P0 A0A1S4H067 A0A182WYP7 B0W1H6 A0A182M1Q6 E9IUX8 A0A1I8N1U1 T1PAE2 A0A182I524 A0A1D2NAU9 A0A0P4Z6L1 A0A0P5R1V6 A0A164ZZF4 A0A0P4ZKJ0 A0A0N7ZML8 A0A224XGU8 A0A0P5YDW3 A0A0P6A9Z7 A0A0N8AEL8 B4LDD9 A0A0P5TMZ2 A0A0N8CYD1 B4L9P7 R4G8X1 A0A146LPN9 A0A226EX63 A0A1A9Y2A5 A0A1B0C6B3 A0A2M3Z972 A0A0P6HW72 A0A0J7L9Y8 A0A1B0GCR2 A0A0P5LJ37 A0A0P5WEK5 A0A0L0CG71 B4IZY8 A0A0P6DXN4
Pubmed
EMBL
KQ460883
KPJ11191.1
AGBW02007871
OWR54458.1
ODYU01012935
SOQ59478.1
+ More
NWSH01000781 PCG74260.1 KQ459603 KPI92769.1 AK401580 BAM18202.1 NNAY01001055 OXU25324.1 NEVH01001351 PNF42940.1 KQ435037 KZC14086.1 QOIP01000003 RLU24990.1 GL444080 EFN61295.1 KK852820 KDR15586.1 KQ981276 KYN43605.1 KZ288269 PBC29971.1 KQ982821 KYQ50175.1 GL453975 EFN75101.1 KQ980249 KYN17099.1 KK107419 EZA51087.1 KQ971343 KYB27165.1 ADTU01004398 ADTU01004399 KQ977004 KYN06502.1 KQ976448 KYM85866.1 GBYB01002456 JAG72223.1 GL887605 EGI70663.1 GEBQ01009943 JAT30034.1 ACPB03015496 GEZM01091073 JAV57119.1 GECZ01026760 GECZ01019840 GECZ01015591 GECZ01005738 JAS43009.1 JAS49929.1 JAS54178.1 JAS64031.1 GEDC01008835 JAS28463.1 JXUM01162821 KQ574611 KXJ67856.1 CVRI01000001 CRK86368.1 CH477514 EAT39720.1 GGFJ01004378 MBW53519.1 GGFJ01004379 MBW53520.1 GGFJ01004377 MBW53518.1 GFDF01001024 JAV13060.1 GFDF01001025 JAV13059.1 AAAB01008984 EAL39051.3 KQ414596 KOC70091.1 ADMH02000797 ETN65037.1 AJVK01007248 GGFK01006911 MBW40232.1 GGFK01006806 MBW40127.1 ATLV01022519 KE525333 KFB47729.1 UFQT01000439 SSX24322.1 GFDL01009871 JAV25174.1 AXCN02000838 GBHO01028624 GBHO01028623 GBHO01006691 GBRD01010308 JAG14980.1 JAG14981.1 JAG36913.1 JAG55516.1 DS231822 EDS25774.1 AXCM01006387 GL766096 EFZ15629.1 KA644898 AFP59527.1 APCN01003750 LJIJ01000113 ODN02384.1 GDIP01219685 JAJ03717.1 GDIQ01106688 JAL45038.1 LRGB01000687 KZS16991.1 GDIP01224581 JAI98820.1 GDIP01230593 JAI92808.1 GFTR01006182 JAW10244.1 GDIP01059203 JAM44512.1 GDIP01032389 JAM71326.1 GDIP01154993 JAJ68409.1 CH940647 EDW68877.1 KRF84059.1 GDIP01124098 JAL79616.1 GDIP01086799 JAM16916.1 CH933816 EDW17095.1 KRG07605.1 GAHY01000232 JAA77278.1 GDHC01011983 GDHC01010042 JAQ06646.1 JAQ08587.1 LNIX01000001 OXA62203.1 JXJN01026528 GGFM01004289 MBW25040.1 GDIQ01013309 JAN81428.1 LBMM01000095 KMR04991.1 CCAG010002190 GDIQ01168878 JAK82847.1 GDIP01087754 JAM15961.1 JRES01000428 KNC31378.1 CH916366 EDV96760.1 GDIQ01070753 JAN23984.1
NWSH01000781 PCG74260.1 KQ459603 KPI92769.1 AK401580 BAM18202.1 NNAY01001055 OXU25324.1 NEVH01001351 PNF42940.1 KQ435037 KZC14086.1 QOIP01000003 RLU24990.1 GL444080 EFN61295.1 KK852820 KDR15586.1 KQ981276 KYN43605.1 KZ288269 PBC29971.1 KQ982821 KYQ50175.1 GL453975 EFN75101.1 KQ980249 KYN17099.1 KK107419 EZA51087.1 KQ971343 KYB27165.1 ADTU01004398 ADTU01004399 KQ977004 KYN06502.1 KQ976448 KYM85866.1 GBYB01002456 JAG72223.1 GL887605 EGI70663.1 GEBQ01009943 JAT30034.1 ACPB03015496 GEZM01091073 JAV57119.1 GECZ01026760 GECZ01019840 GECZ01015591 GECZ01005738 JAS43009.1 JAS49929.1 JAS54178.1 JAS64031.1 GEDC01008835 JAS28463.1 JXUM01162821 KQ574611 KXJ67856.1 CVRI01000001 CRK86368.1 CH477514 EAT39720.1 GGFJ01004378 MBW53519.1 GGFJ01004379 MBW53520.1 GGFJ01004377 MBW53518.1 GFDF01001024 JAV13060.1 GFDF01001025 JAV13059.1 AAAB01008984 EAL39051.3 KQ414596 KOC70091.1 ADMH02000797 ETN65037.1 AJVK01007248 GGFK01006911 MBW40232.1 GGFK01006806 MBW40127.1 ATLV01022519 KE525333 KFB47729.1 UFQT01000439 SSX24322.1 GFDL01009871 JAV25174.1 AXCN02000838 GBHO01028624 GBHO01028623 GBHO01006691 GBRD01010308 JAG14980.1 JAG14981.1 JAG36913.1 JAG55516.1 DS231822 EDS25774.1 AXCM01006387 GL766096 EFZ15629.1 KA644898 AFP59527.1 APCN01003750 LJIJ01000113 ODN02384.1 GDIP01219685 JAJ03717.1 GDIQ01106688 JAL45038.1 LRGB01000687 KZS16991.1 GDIP01224581 JAI98820.1 GDIP01230593 JAI92808.1 GFTR01006182 JAW10244.1 GDIP01059203 JAM44512.1 GDIP01032389 JAM71326.1 GDIP01154993 JAJ68409.1 CH940647 EDW68877.1 KRF84059.1 GDIP01124098 JAL79616.1 GDIP01086799 JAM16916.1 CH933816 EDW17095.1 KRG07605.1 GAHY01000232 JAA77278.1 GDHC01011983 GDHC01010042 JAQ06646.1 JAQ08587.1 LNIX01000001 OXA62203.1 JXJN01026528 GGFM01004289 MBW25040.1 GDIQ01013309 JAN81428.1 LBMM01000095 KMR04991.1 CCAG010002190 GDIQ01168878 JAK82847.1 GDIP01087754 JAM15961.1 JRES01000428 KNC31378.1 CH916366 EDV96760.1 GDIQ01070753 JAN23984.1
Proteomes
UP000053240
UP000007151
UP000218220
UP000053268
UP000002358
UP000215335
+ More
UP000235965 UP000076502 UP000279307 UP000000311 UP000027135 UP000078541 UP000005203 UP000242457 UP000075809 UP000008237 UP000078492 UP000053097 UP000007266 UP000005205 UP000078542 UP000192223 UP000078540 UP000007755 UP000015103 UP000069940 UP000249989 UP000183832 UP000008820 UP000075884 UP000007062 UP000075901 UP000076408 UP000053825 UP000000673 UP000075920 UP000075903 UP000092462 UP000030765 UP000075900 UP000069272 UP000075886 UP000075880 UP000075882 UP000075885 UP000075902 UP000076407 UP000002320 UP000075883 UP000095301 UP000075840 UP000094527 UP000076858 UP000008792 UP000009192 UP000198287 UP000092443 UP000092460 UP000036403 UP000092444 UP000037069 UP000001070
UP000235965 UP000076502 UP000279307 UP000000311 UP000027135 UP000078541 UP000005203 UP000242457 UP000075809 UP000008237 UP000078492 UP000053097 UP000007266 UP000005205 UP000078542 UP000192223 UP000078540 UP000007755 UP000015103 UP000069940 UP000249989 UP000183832 UP000008820 UP000075884 UP000007062 UP000075901 UP000076408 UP000053825 UP000000673 UP000075920 UP000075903 UP000092462 UP000030765 UP000075900 UP000069272 UP000075886 UP000075880 UP000075882 UP000075885 UP000075902 UP000076407 UP000002320 UP000075883 UP000095301 UP000075840 UP000094527 UP000076858 UP000008792 UP000009192 UP000198287 UP000092443 UP000092460 UP000036403 UP000092444 UP000037069 UP000001070
ProteinModelPortal
A0A194R5Q0
A0A212FL33
A0A2H1X2K8
A0A2A4JS76
A0A194PJ71
I4DJW2
+ More
K7J0J1 A0A232F3I7 A0A2J7RQ39 A0A154PQ94 A0A3L8DWZ2 E2AZ37 A0A067QYM4 A0A195FSN7 A0A087ZYU2 A0A2A3EG43 A0A151WR73 E2CAD5 A0A151J3T4 A0A026W4U8 A0A139WH37 A0A158NYY2 A0A195D0W2 A0A1W4WSN9 A0A151I4P8 A0A0C9R6R8 F4W567 A0A1B6M2B5 T1HXI5 A0A1Y1K6N3 A0A1B6EYD2 A0A1B6DS34 A0A182GEQ5 A0A1J1HEH2 Q16YL5 A0A1S4FJS7 A0A2M4BKB5 A0A2M4BKD3 A0A2M4BL53 A0A1L8E2U1 A0A182NF04 A0A1L8E344 Q5TND9 A0A182STQ0 A0A182YFB5 A0A0L7RH96 W5JKY5 A0A182WH27 A0A182V7F0 A0A1B0GQI2 A0A2M4AHN4 A0A2M4AHB0 A0A084WBY5 A0A336M257 A0A1Q3FC71 A0A1Y9HEF4 A0A182F5D6 A0A182Q5V3 A0A182JF51 A0A182KUL8 A0A182NZW3 A0A182TZY2 A0A0A9Z0P0 A0A1S4H067 A0A182WYP7 B0W1H6 A0A182M1Q6 E9IUX8 A0A1I8N1U1 T1PAE2 A0A182I524 A0A1D2NAU9 A0A0P4Z6L1 A0A0P5R1V6 A0A164ZZF4 A0A0P4ZKJ0 A0A0N7ZML8 A0A224XGU8 A0A0P5YDW3 A0A0P6A9Z7 A0A0N8AEL8 B4LDD9 A0A0P5TMZ2 A0A0N8CYD1 B4L9P7 R4G8X1 A0A146LPN9 A0A226EX63 A0A1A9Y2A5 A0A1B0C6B3 A0A2M3Z972 A0A0P6HW72 A0A0J7L9Y8 A0A1B0GCR2 A0A0P5LJ37 A0A0P5WEK5 A0A0L0CG71 B4IZY8 A0A0P6DXN4
K7J0J1 A0A232F3I7 A0A2J7RQ39 A0A154PQ94 A0A3L8DWZ2 E2AZ37 A0A067QYM4 A0A195FSN7 A0A087ZYU2 A0A2A3EG43 A0A151WR73 E2CAD5 A0A151J3T4 A0A026W4U8 A0A139WH37 A0A158NYY2 A0A195D0W2 A0A1W4WSN9 A0A151I4P8 A0A0C9R6R8 F4W567 A0A1B6M2B5 T1HXI5 A0A1Y1K6N3 A0A1B6EYD2 A0A1B6DS34 A0A182GEQ5 A0A1J1HEH2 Q16YL5 A0A1S4FJS7 A0A2M4BKB5 A0A2M4BKD3 A0A2M4BL53 A0A1L8E2U1 A0A182NF04 A0A1L8E344 Q5TND9 A0A182STQ0 A0A182YFB5 A0A0L7RH96 W5JKY5 A0A182WH27 A0A182V7F0 A0A1B0GQI2 A0A2M4AHN4 A0A2M4AHB0 A0A084WBY5 A0A336M257 A0A1Q3FC71 A0A1Y9HEF4 A0A182F5D6 A0A182Q5V3 A0A182JF51 A0A182KUL8 A0A182NZW3 A0A182TZY2 A0A0A9Z0P0 A0A1S4H067 A0A182WYP7 B0W1H6 A0A182M1Q6 E9IUX8 A0A1I8N1U1 T1PAE2 A0A182I524 A0A1D2NAU9 A0A0P4Z6L1 A0A0P5R1V6 A0A164ZZF4 A0A0P4ZKJ0 A0A0N7ZML8 A0A224XGU8 A0A0P5YDW3 A0A0P6A9Z7 A0A0N8AEL8 B4LDD9 A0A0P5TMZ2 A0A0N8CYD1 B4L9P7 R4G8X1 A0A146LPN9 A0A226EX63 A0A1A9Y2A5 A0A1B0C6B3 A0A2M3Z972 A0A0P6HW72 A0A0J7L9Y8 A0A1B0GCR2 A0A0P5LJ37 A0A0P5WEK5 A0A0L0CG71 B4IZY8 A0A0P6DXN4
Ontologies
PANTHER
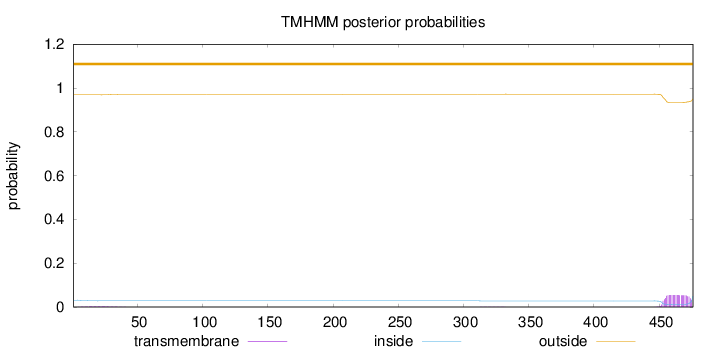
Topology
Length:
476
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21774
Exp number, first 60 AAs:
0.06482
Total prob of N-in:
0.03119
outside
1 - 476
Population Genetic Test Statistics
Pi
156.417766
Theta
145.071798
Tajima's D
-0.852006
CLR
1.054491
CSRT
0.165891705414729
Interpretation
Uncertain
