Gene
KWMTBOMO07196
Pre Gene Modal
BGIBMGA010384
Annotation
PREDICTED:_decapentaplegic_isoform_X1_[Bombyx_mori]
Full name
Protein decapentaplegic
Location in the cell
Mitochondrial Reliability : 1.555 Nuclear Reliability : 1.46
Sequence
CDS
ATGCGTGGGGCGTGCGCATGCGCGGTGGTGTGCGCGTTGGTGGCGCTGTGCGCAGCCGCGGGCCTCGACGAAGCGACGCGTGTTGCTGCAGAGAAGCAGTTATTGGCGTTGTTGGGTCTGCCGAAGAGACCGTCGCGTCGATCCGCTCCAGTACCGCCTATACCACGCGCCATGCGAATGCTTTACGAGGCAAGCGGAGCCATACCGGCCGCTGCGGCAAACACGGCCCGTTCATATCAGCACGTACCGACGGAGCTCGATGCGAGGTTCCCAGGCGAACATCGTTTTCGCCTATTCTTCAACCTAAGTGGAGTACCCTCTGATGAAGTAGCTCGTGGTGCTGATCTCAAATTTCATCGCGCGACTGAAGAGACGGGTCCTCAGCGCCTATTACTATATGACGTTGTACGTCCTGGTCGTCGAGGGAAAACGACTCCAATTCTAAGACTTCTCGATTCCGTGACATTATTGCCAGGCGAGGGCACAGTGACAGCAGACGCCATTGACGCAGTGCGACGGTGGCTCCTTGAAACTGATCAAAACCATGGACTATTAGTGCGTGTTATTGAAGAAGGCCAACACAACGTTGATGCAAAACGGCCACACGTAAGAGTTCGAAGACGAGCGACCGAAGACGAAGAAGAATGGCGCTCTCAGCAACCCTTGCTGTTGCTGTACACTGAAGATGCGCGAGCCAGAGAAGCACGCGAGAATGGGGAGTCGCGTCTAACTCGAAATAAAAGAGCAACACAACGGCGTGGTCATCGACCTCACCACCGTCGTAAAGAAGCTCGTGAAATCTGCCAGCGGCGGCCACTGTTTGTCGACTTCGCGGAAGTCGGCTGGAGCGACTGGATTGTCGCGCCTCCTGGTTACGAAGCTTATTTCTGTCAGGGTGACTGCCCGTTCCCGCTCGCAGATCATCTAAATGGCACTAATCACGCAATAGTGCAAACTTTAGTGAATTCAGTGGACCCAGCCTTAGTGCCTAAAGCGTGTTGTATACCAACACAACTATCGCCTATTTCTATGTTATATATGGACGAACATAATCAAGTGGTGCTTAAAAACTATCAGGATATGATGGTGATGGGTTGCGGTTGCCGATGA
Protein
MRGACACAVVCALVALCAAAGLDEATRVAAEKQLLALLGLPKRPSRRSAPVPPIPRAMRMLYEASGAIPAAAANTARSYQHVPTELDARFPGEHRFRLFFNLSGVPSDEVARGADLKFHRATEETGPQRLLLYDVVRPGRRGKTTPILRLLDSVTLLPGEGTVTADAIDAVRRWLLETDQNHGLLVRVIEEGQHNVDAKRPHVRVRRRATEDEEEWRSQQPLLLLYTEDARAREARENGESRLTRNKRATQRRGHRPHHRRKEAREICQRRPLFVDFAEVGWSDWIVAPPGYEAYFCQGDCPFPLADHLNGTNHAIVQTLVNSVDPALVPKACCIPTQLSPISMLYMDEHNQVVLKNYQDMMVMGCGCR
Summary
Description
Required during oogenesis for eggshell patterning and dorsal/ventral patterning of the embryo. Acts as a morphogen during embryogenesis to pattern the dorsal/ventral axis, specifying dorsal ectoderm and amnioserosa cell fate within the dorsal half of the embryo; this activity is antagonized by binding to sog and tsg. Induces the formation of visceral mesoderm and the heart in early embryos. Required later in embryogenesis for dorsal closure and patterning of the hindgut. Also functions postembryonically as a long-range morphogen during imaginal disk development; is responsible for the progression of the morphogenetic furrow during eye development. Patterns the wing imaginal disk along its anterior/posterior axis and has a role in positioning pro-veins. Also required to subdivide the wing disk along the proximal/distal axis into body wall (notum) and wing. Ensures the correct architecture of wing epithelial cells. Has multiple roles in the developing tracheal system, controlling directed tracheal cell migration during embryogenesis and later specifying the fate of fusion cells in the tracheal branches. Required for viability of larvae. Essential for the maintenance and division of germline stem cells in the ovary. Signals via the type I receptor tkv, the type II receptor punt, and in some tissues via the type I receptor sax, in a signaling cascade that leads to activation and repression of target genes.
Acts as an extracellular morphogen to establish at least two cellular response thresholds within the dorsal half of the drosophila embryo. Required for the proper development of the embryonic dorsal hypoderm, for viability of larvae and for cell viability of the epithelial cells in the imaginal disks. Acts together with scw (By similarity).
Acts as an extracellular morphogen to establish at least two cellular response thresholds within the dorsal half of the drosophila embryo. Required for the proper development of the embryonic dorsal hypoderm, for viability of larvae and for cell viability of the epithelial cells in the imaginal disks. Acts together with scw (By similarity).
Subunit
Heterodimers of scw/dpp are the active subunit, dpp/dpp homodimers elicit a basal response and scw/scw homodimers alone are ineffective in specifying a dorsal pattern. Component of a complex composed of dpp, sog and tsg.
Heterodimers of scw/dpp are the active subunit, dpp/dpp homodimers elicit a basal response and scw/scw homodimers alone are ineffective in specifying a dorsal pattern.
Heterodimers of scw/dpp are the active subunit, dpp/dpp homodimers elicit a basal response and scw/scw homodimers alone are ineffective in specifying a dorsal pattern.
Similarity
Belongs to the TGF-beta family.
Keywords
Complete proteome
Developmental protein
Differentiation
Direct protein sequencing
Disulfide bond
Glycoprotein
Growth factor
Polymorphism
Reference proteome
Secreted
Signal
Feature
chain Protein decapentaplegic
Uniprot
H9JLI3
A0A0F7IQS1
B8YPW1
A0A194PHI0
A0A2A4JRC3
A0A212FL92
+ More
A0A2H1WNA7 A0A194R100 E3UKJ8 A0A1B0DI00 A0A1B0CNU7 A0A1L8DL37 A0A1L8DLA2 A0A1L8DJC3 A0A2A4JR18 A0A1B6MG22 A0A1S4F007 A0A1J1IM20 Q17JZ3 A0A182GD36 Q75WK6 A0A1S4JCR2 B0WCI2 A0A182NTL6 A0A2J7Q4Z8 A0A182J5G4 A0A182XZW8 A0A182K0K3 A0A182QNM0 Q7Q3Q7 A0A0C9RHG2 A0A182P4R0 A0A2P8YVQ1 A0A336LZP6 A0A182L1W9 A0A182RFJ7 A0A182TTR3 A0A182F1K1 W5JTS3 A0A1B6DZH3 A0A182UWV4 A0A182HV19 A0A182WT22 A0A1S4GXL4 A0A151XAT1 E2B9W4 A0A151J8N4 A0A195BSU4 A0A158P1S4 I3WD96 W8BL12 A0A0K8WG48 F4W6N8 E2AGL6 A0A2M4CGV9 A0A195ERI5 A0A182W7F5 A0A0K8U826 A0A3L8DG15 A0A026W2E0 A0A034VC06 A0A195CMC5 A0A1Y1LN98 A0A0A1X980 A0A0L0C8W2 A0A154P4L3 K7IMN3 A0A140AX77 A0A0L7R784 V9IGS0 A0A140AX76 A0A1W4WXB0 A0A1I8P6N3 T1PJE3 A0A1S3DKS8 A0A1B0REL8 T1H8Q4 A0A224XSA9 A0A1I8MHF7 A0A290Y240 A0A0N0BCB8 B4MU02 A0A023F2N8 B4KGU4 B4Q848 A0A182LWX6 A0A0M3QT42 B3NA13 P07713 P91706 Q1WKY7 Q1WKY8 B4NWQ1 Q1WKY6 A0A0Q5W8W8 A0A088AJM8 A0A0R1DRG4 A0A067YM92 D0QWH2
A0A2H1WNA7 A0A194R100 E3UKJ8 A0A1B0DI00 A0A1B0CNU7 A0A1L8DL37 A0A1L8DLA2 A0A1L8DJC3 A0A2A4JR18 A0A1B6MG22 A0A1S4F007 A0A1J1IM20 Q17JZ3 A0A182GD36 Q75WK6 A0A1S4JCR2 B0WCI2 A0A182NTL6 A0A2J7Q4Z8 A0A182J5G4 A0A182XZW8 A0A182K0K3 A0A182QNM0 Q7Q3Q7 A0A0C9RHG2 A0A182P4R0 A0A2P8YVQ1 A0A336LZP6 A0A182L1W9 A0A182RFJ7 A0A182TTR3 A0A182F1K1 W5JTS3 A0A1B6DZH3 A0A182UWV4 A0A182HV19 A0A182WT22 A0A1S4GXL4 A0A151XAT1 E2B9W4 A0A151J8N4 A0A195BSU4 A0A158P1S4 I3WD96 W8BL12 A0A0K8WG48 F4W6N8 E2AGL6 A0A2M4CGV9 A0A195ERI5 A0A182W7F5 A0A0K8U826 A0A3L8DG15 A0A026W2E0 A0A034VC06 A0A195CMC5 A0A1Y1LN98 A0A0A1X980 A0A0L0C8W2 A0A154P4L3 K7IMN3 A0A140AX77 A0A0L7R784 V9IGS0 A0A140AX76 A0A1W4WXB0 A0A1I8P6N3 T1PJE3 A0A1S3DKS8 A0A1B0REL8 T1H8Q4 A0A224XSA9 A0A1I8MHF7 A0A290Y240 A0A0N0BCB8 B4MU02 A0A023F2N8 B4KGU4 B4Q848 A0A182LWX6 A0A0M3QT42 B3NA13 P07713 P91706 Q1WKY7 Q1WKY8 B4NWQ1 Q1WKY6 A0A0Q5W8W8 A0A088AJM8 A0A0R1DRG4 A0A067YM92 D0QWH2
Pubmed
19121390
26354079
22118469
17510324
26483478
14872342
+ More
25244985 12364791 15302159 14747013 17210077 29403074 20966253 20920257 23761445 20798317 21347285 24495485 21719571 30249741 24508170 25348373 28004739 25830018 26108605 20075255 26881804 25315136 17994087 25474469 18057021 3467201 9071586 10731132 12537572 12618399 1692958 3123323 8252627 7720583 7700357 9226445 9695953 9636086 10495279 11136981 11136982 10903186 11432817 11231061 11260716 12135920 15797386 15867930 9071585 16387879 17550304 24886828 19859648
25244985 12364791 15302159 14747013 17210077 29403074 20966253 20920257 23761445 20798317 21347285 24495485 21719571 30249741 24508170 25348373 28004739 25830018 26108605 20075255 26881804 25315136 17994087 25474469 18057021 3467201 9071586 10731132 12537572 12618399 1692958 3123323 8252627 7720583 7700357 9226445 9695953 9636086 10495279 11136981 11136982 10903186 11432817 11231061 11260716 12135920 15797386 15867930 9071585 16387879 17550304 24886828 19859648
EMBL
BABH01005698
KP861420
AKG96797.1
FJ572058
ACL97675.1
KQ459603
+ More
KPI92772.1 NWSH01000781 PCG74258.1 AGBW02007871 OWR54459.1 ODYU01009769 SOQ54422.1 KQ460883 KPJ11189.1 HM449872 ADO33007.1 AJVK01006118 AJWK01020933 GFDF01006908 JAV07176.1 GFDF01006862 JAV07222.1 GFDF01007617 JAV06467.1 PCG74259.1 GEBQ01005121 JAT34856.1 CVRI01000055 CRL01208.1 CH477229 EAT47007.1 JXUM01055189 JXUM01055190 JXUM01055191 KQ561855 KXJ77311.1 AB121072 BAD08319.1 DS231888 EDS43553.1 NEVH01018372 PNF23658.1 AXCN02000795 AY578800 AAAB01008964 AAT07305.1 EAA12482.2 GBYB01012642 JAG82409.1 PYGN01000334 PSN48329.1 UFQT01000238 SSX22219.1 ADMH02000511 ETN66149.1 GEDC01006216 GEDC01005432 JAS31082.1 JAS31866.1 APCN01002490 KQ982335 KYQ57449.1 GL446605 EFN87551.1 KQ979492 KYN21220.1 KQ976419 KYM89365.1 ADTU01006778 ADTU01006779 JX021498 AFL02791.1 GAMC01008877 JAB97678.1 GDHF01002499 JAI49815.1 GL887707 EGI70114.1 GL439322 EFN67436.1 GGFL01000366 MBW64544.1 KQ982021 KYN30489.1 GDHF01029591 JAI22723.1 QOIP01000008 RLU19395.1 KK107536 EZA49199.1 GAKP01019652 JAC39300.1 KQ977595 KYN01622.1 GEZM01052697 GEZM01052695 JAV74318.1 GBXI01015540 GBXI01006488 JAC98751.1 JAD07804.1 JRES01000753 KNC28677.1 KQ434816 KZC06856.1 KT750953 ALG65084.1 KQ414642 KOC66742.1 JR046821 AEY60260.1 KT750952 KZ288349 ALG65083.1 PBC27446.1 KA648814 AFP63443.1 KM264380 AKE98360.1 ACPB03020299 GFTR01005403 JAW11023.1 KX951674 ATD85058.1 KQ435916 KOX68761.1 CH963852 EDW75591.1 GBBI01003269 JAC15443.1 CH933807 EDW11144.2 KRG02541.1 CM000361 EDX03470.1 AXCM01006054 CP012523 ALC38229.1 CH954177 EDV57476.1 KQS70002.1 M30116 U63857 AE014134 BT015227 AF459545 AF459546 AF459547 AF459548 AF459549 AF459550 AF459551 AF459552 AF459553 AF459554 AF459555 AF459556 AF459557 AF459558 AF459559 AF459560 AF459561 AF459562 AF459563 AF459564 U63854 AH015352 ABB29567.1 AH015351 ABB29566.1 CM000157 EDW87393.2 AH015353 DQ167739 ABB29568.1 KQS70001.1 KRJ97142.1 KF630593 AHC98624.1 FJ821028 ACN94671.1
KPI92772.1 NWSH01000781 PCG74258.1 AGBW02007871 OWR54459.1 ODYU01009769 SOQ54422.1 KQ460883 KPJ11189.1 HM449872 ADO33007.1 AJVK01006118 AJWK01020933 GFDF01006908 JAV07176.1 GFDF01006862 JAV07222.1 GFDF01007617 JAV06467.1 PCG74259.1 GEBQ01005121 JAT34856.1 CVRI01000055 CRL01208.1 CH477229 EAT47007.1 JXUM01055189 JXUM01055190 JXUM01055191 KQ561855 KXJ77311.1 AB121072 BAD08319.1 DS231888 EDS43553.1 NEVH01018372 PNF23658.1 AXCN02000795 AY578800 AAAB01008964 AAT07305.1 EAA12482.2 GBYB01012642 JAG82409.1 PYGN01000334 PSN48329.1 UFQT01000238 SSX22219.1 ADMH02000511 ETN66149.1 GEDC01006216 GEDC01005432 JAS31082.1 JAS31866.1 APCN01002490 KQ982335 KYQ57449.1 GL446605 EFN87551.1 KQ979492 KYN21220.1 KQ976419 KYM89365.1 ADTU01006778 ADTU01006779 JX021498 AFL02791.1 GAMC01008877 JAB97678.1 GDHF01002499 JAI49815.1 GL887707 EGI70114.1 GL439322 EFN67436.1 GGFL01000366 MBW64544.1 KQ982021 KYN30489.1 GDHF01029591 JAI22723.1 QOIP01000008 RLU19395.1 KK107536 EZA49199.1 GAKP01019652 JAC39300.1 KQ977595 KYN01622.1 GEZM01052697 GEZM01052695 JAV74318.1 GBXI01015540 GBXI01006488 JAC98751.1 JAD07804.1 JRES01000753 KNC28677.1 KQ434816 KZC06856.1 KT750953 ALG65084.1 KQ414642 KOC66742.1 JR046821 AEY60260.1 KT750952 KZ288349 ALG65083.1 PBC27446.1 KA648814 AFP63443.1 KM264380 AKE98360.1 ACPB03020299 GFTR01005403 JAW11023.1 KX951674 ATD85058.1 KQ435916 KOX68761.1 CH963852 EDW75591.1 GBBI01003269 JAC15443.1 CH933807 EDW11144.2 KRG02541.1 CM000361 EDX03470.1 AXCM01006054 CP012523 ALC38229.1 CH954177 EDV57476.1 KQS70002.1 M30116 U63857 AE014134 BT015227 AF459545 AF459546 AF459547 AF459548 AF459549 AF459550 AF459551 AF459552 AF459553 AF459554 AF459555 AF459556 AF459557 AF459558 AF459559 AF459560 AF459561 AF459562 AF459563 AF459564 U63854 AH015352 ABB29567.1 AH015351 ABB29566.1 CM000157 EDW87393.2 AH015353 DQ167739 ABB29568.1 KQS70001.1 KRJ97142.1 KF630593 AHC98624.1 FJ821028 ACN94671.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000092462
+ More
UP000092461 UP000183832 UP000008820 UP000069940 UP000249989 UP000002320 UP000075884 UP000235965 UP000075880 UP000076408 UP000075881 UP000075886 UP000007062 UP000075885 UP000245037 UP000075882 UP000075900 UP000075902 UP000069272 UP000000673 UP000075903 UP000075840 UP000076407 UP000075809 UP000008237 UP000078492 UP000078540 UP000005205 UP000007755 UP000000311 UP000078541 UP000075920 UP000279307 UP000053097 UP000078542 UP000037069 UP000076502 UP000002358 UP000053825 UP000242457 UP000192223 UP000095300 UP000079169 UP000015103 UP000095301 UP000053105 UP000007798 UP000009192 UP000000304 UP000075883 UP000092553 UP000008711 UP000000803 UP000002282 UP000005203
UP000092461 UP000183832 UP000008820 UP000069940 UP000249989 UP000002320 UP000075884 UP000235965 UP000075880 UP000076408 UP000075881 UP000075886 UP000007062 UP000075885 UP000245037 UP000075882 UP000075900 UP000075902 UP000069272 UP000000673 UP000075903 UP000075840 UP000076407 UP000075809 UP000008237 UP000078492 UP000078540 UP000005205 UP000007755 UP000000311 UP000078541 UP000075920 UP000279307 UP000053097 UP000078542 UP000037069 UP000076502 UP000002358 UP000053825 UP000242457 UP000192223 UP000095300 UP000079169 UP000015103 UP000095301 UP000053105 UP000007798 UP000009192 UP000000304 UP000075883 UP000092553 UP000008711 UP000000803 UP000002282 UP000005203
Interpro
Gene 3D
ProteinModelPortal
H9JLI3
A0A0F7IQS1
B8YPW1
A0A194PHI0
A0A2A4JRC3
A0A212FL92
+ More
A0A2H1WNA7 A0A194R100 E3UKJ8 A0A1B0DI00 A0A1B0CNU7 A0A1L8DL37 A0A1L8DLA2 A0A1L8DJC3 A0A2A4JR18 A0A1B6MG22 A0A1S4F007 A0A1J1IM20 Q17JZ3 A0A182GD36 Q75WK6 A0A1S4JCR2 B0WCI2 A0A182NTL6 A0A2J7Q4Z8 A0A182J5G4 A0A182XZW8 A0A182K0K3 A0A182QNM0 Q7Q3Q7 A0A0C9RHG2 A0A182P4R0 A0A2P8YVQ1 A0A336LZP6 A0A182L1W9 A0A182RFJ7 A0A182TTR3 A0A182F1K1 W5JTS3 A0A1B6DZH3 A0A182UWV4 A0A182HV19 A0A182WT22 A0A1S4GXL4 A0A151XAT1 E2B9W4 A0A151J8N4 A0A195BSU4 A0A158P1S4 I3WD96 W8BL12 A0A0K8WG48 F4W6N8 E2AGL6 A0A2M4CGV9 A0A195ERI5 A0A182W7F5 A0A0K8U826 A0A3L8DG15 A0A026W2E0 A0A034VC06 A0A195CMC5 A0A1Y1LN98 A0A0A1X980 A0A0L0C8W2 A0A154P4L3 K7IMN3 A0A140AX77 A0A0L7R784 V9IGS0 A0A140AX76 A0A1W4WXB0 A0A1I8P6N3 T1PJE3 A0A1S3DKS8 A0A1B0REL8 T1H8Q4 A0A224XSA9 A0A1I8MHF7 A0A290Y240 A0A0N0BCB8 B4MU02 A0A023F2N8 B4KGU4 B4Q848 A0A182LWX6 A0A0M3QT42 B3NA13 P07713 P91706 Q1WKY7 Q1WKY8 B4NWQ1 Q1WKY6 A0A0Q5W8W8 A0A088AJM8 A0A0R1DRG4 A0A067YM92 D0QWH2
A0A2H1WNA7 A0A194R100 E3UKJ8 A0A1B0DI00 A0A1B0CNU7 A0A1L8DL37 A0A1L8DLA2 A0A1L8DJC3 A0A2A4JR18 A0A1B6MG22 A0A1S4F007 A0A1J1IM20 Q17JZ3 A0A182GD36 Q75WK6 A0A1S4JCR2 B0WCI2 A0A182NTL6 A0A2J7Q4Z8 A0A182J5G4 A0A182XZW8 A0A182K0K3 A0A182QNM0 Q7Q3Q7 A0A0C9RHG2 A0A182P4R0 A0A2P8YVQ1 A0A336LZP6 A0A182L1W9 A0A182RFJ7 A0A182TTR3 A0A182F1K1 W5JTS3 A0A1B6DZH3 A0A182UWV4 A0A182HV19 A0A182WT22 A0A1S4GXL4 A0A151XAT1 E2B9W4 A0A151J8N4 A0A195BSU4 A0A158P1S4 I3WD96 W8BL12 A0A0K8WG48 F4W6N8 E2AGL6 A0A2M4CGV9 A0A195ERI5 A0A182W7F5 A0A0K8U826 A0A3L8DG15 A0A026W2E0 A0A034VC06 A0A195CMC5 A0A1Y1LN98 A0A0A1X980 A0A0L0C8W2 A0A154P4L3 K7IMN3 A0A140AX77 A0A0L7R784 V9IGS0 A0A140AX76 A0A1W4WXB0 A0A1I8P6N3 T1PJE3 A0A1S3DKS8 A0A1B0REL8 T1H8Q4 A0A224XSA9 A0A1I8MHF7 A0A290Y240 A0A0N0BCB8 B4MU02 A0A023F2N8 B4KGU4 B4Q848 A0A182LWX6 A0A0M3QT42 B3NA13 P07713 P91706 Q1WKY7 Q1WKY8 B4NWQ1 Q1WKY6 A0A0Q5W8W8 A0A088AJM8 A0A0R1DRG4 A0A067YM92 D0QWH2
PDB
2QJB
E-value=3.32734e-48,
Score=483
Ontologies
PATHWAY
GO
GO:0005576
GO:0008083
GO:0016021
GO:0043408
GO:0005615
GO:0042981
GO:0030509
GO:0060395
GO:0005160
GO:0048468
GO:0010862
GO:0005125
GO:0008354
GO:0007458
GO:0070700
GO:0098898
GO:0036099
GO:0035263
GO:0061327
GO:0007498
GO:0035217
GO:0007516
GO:0007352
GO:0036098
GO:0001715
GO:0035224
GO:0035222
GO:0061320
GO:0007427
GO:0061352
GO:0035156
GO:0030721
GO:0045595
GO:0048636
GO:0008201
GO:0046982
GO:0060323
GO:0035158
GO:0042803
GO:0031252
GO:0045572
GO:0007391
GO:0007474
GO:0016015
GO:0035230
GO:0010002
GO:0007455
GO:0005768
GO:0010629
GO:0042078
GO:0007313
GO:0008586
GO:0005518
GO:0035168
GO:0007378
GO:0035309
GO:0046843
GO:0010628
GO:0001708
GO:0001745
GO:0007304
GO:0007440
GO:0008360
GO:0048619
GO:0007448
GO:0001709
GO:0035215
GO:0019827
GO:0048100
GO:0045705
GO:0007479
GO:0008285
GO:0007423
GO:0030718
GO:0045476
GO:0048542
GO:0007476
GO:0007446
GO:0007354
GO:0048066
GO:0009950
GO:0007447
GO:0030707
GO:0007393
GO:0007442
GO:0007179
GO:0009948
GO:0007443
GO:0007507
GO:0007444
GO:0042127
GO:0007473
GO:0007424
GO:0007398
GO:0045570
GO:0009953
GO:0007281
GO:0035214
GO:0007275
GO:0030154
GO:0040007
GO:0016705
GO:0031418
GO:0016491
GO:0004656
GO:0005783
GO:0016702
GO:0016295
GO:0016296
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 19,
Likelihood: 0.966154
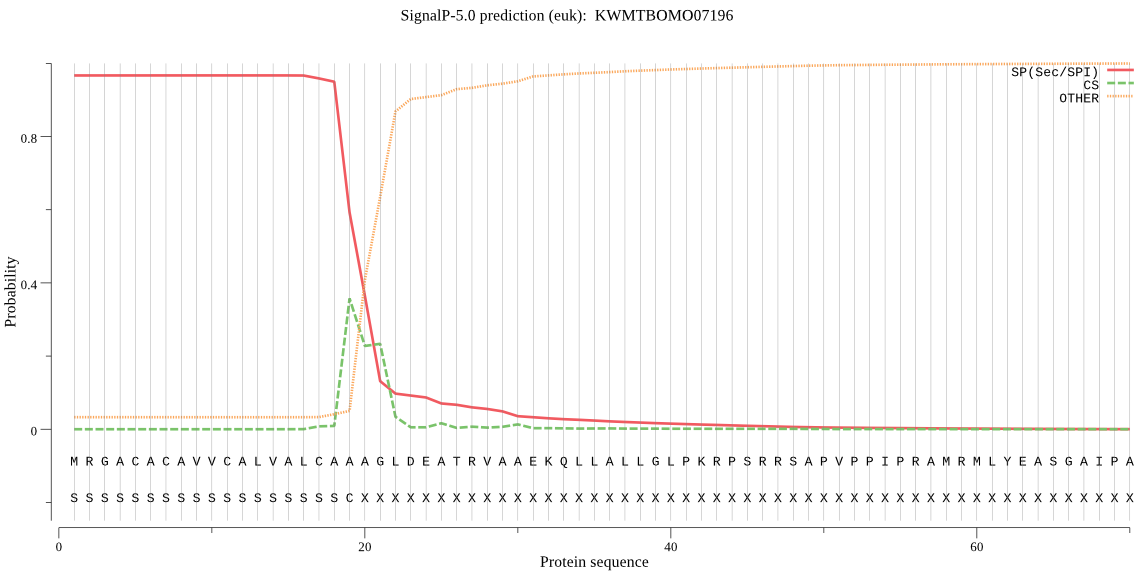
Length:
369
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.02578
Exp number, first 60 AAs:
10.02251
Total prob of N-in:
0.51713
POSSIBLE N-term signal
sequence
outside
1 - 369
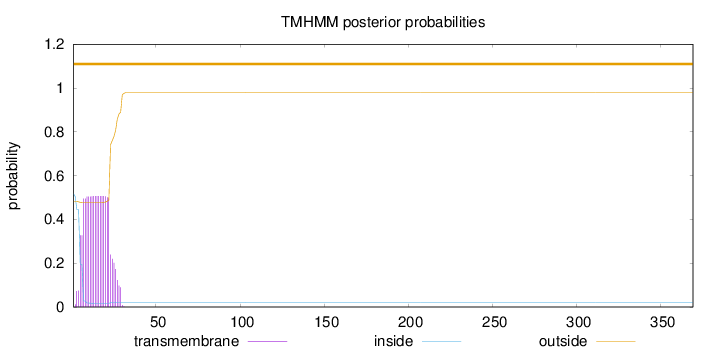
Population Genetic Test Statistics
Pi
45.490727
Theta
45.835786
Tajima's D
-1.083911
CLR
27.097602
CSRT
0.124943752812359
Interpretation
Uncertain
