Gene
KWMTBOMO07195 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010569
Annotation
PREDICTED:_39S_ribosomal_protein_L46?_mitochondrial_[Amyelois_transitella]
Full name
Exocyst complex component 7
Alternative Name
Exocyst complex component Exo70
Location in the cell
Cytoplasmic Reliability : 1.092 Extracellular Reliability : 1.144 Nuclear Reliability : 1.655
Sequence
CDS
ATGTTGGTTAAGACATTGATGCGTATGAATATAACTTATTACCACAGGCTTTTAATAAGAAGTATTAGTAGCAAGCCAGCTTGGGATATATTGACGGGTATTTGCATTGAACGGTTACCAGTCGTAACTCCGCCGTTAACTGAAATGCAAAAAAAATATAAAGAATTTTTATTGACCGTAGAATTTGAGAAAAGCATGAAATCAAACCATGAAATTCAACACGAAAATGACAAAAAACAAGCTGAACTTTTCAAGGCTGAATCCCAAGATGTTGATATTGATGCTGTTAATAAAGTTACCGCCCAAGATTTTGAAGATGCAGCAAATGAAGAGTATTCAAAATTTAAATTTGGAAATTTAGAAACAGATGCAGACAAAAAAGGTGACAAAACATCAACAGAACGGTGCTTACAACGACATCTAGTTCTTGTGACACAACGGAAATTAGGAAATGATTCAAAGACATTACTTCCTCAGGGGCATTGGCAGGAGGGCGAGACACTACGTCAGACTGCTGAAAGAATAGTTAAGGAGCAAATCGGATCTGAACTTCAGATAAAATTTATTTCAAATGCTCCATGTGGATTCTACAAATATAAATATCCCAGTGAAATGAATGGAAAAGTTGGCGCCAAAATTTTTTTCTACTATGCAAACTACAAAAGTGGCAATCCTACAAAGTCTAAAGTGAATTGGTTAACACGTAAAGAACTCGATGAAATGTTACCACAGAAATATAGTAATGCAGTCAAAGAATTTTTAATTGAAGAAACTTATTAA
Protein
MLVKTLMRMNITYYHRLLIRSISSKPAWDILTGICIERLPVVTPPLTEMQKKYKEFLLTVEFEKSMKSNHEIQHENDKKQAELFKAESQDVDIDAVNKVTAQDFEDAANEEYSKFKFGNLETDADKKGDKTSTERCLQRHLVLVTQRKLGNDSKTLLPQGHWQEGETLRQTAERIVKEQIGSELQIKFISNAPCGFYKYKYPSEMNGKVGAKIFFYYANYKSGNPTKSKVNWLTRKELDEMLPQKYSNAVKEFLIEETY
Summary
Description
Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane.
Similarity
Belongs to the EXO70 family.
Uniprot
H9JM17
A0A2H1W792
A0A2A4JQB3
A0A0N1I7W3
I4DNZ4
A0A194PIA1
+ More
A0A212FL90 A0A194R061 A0A1Y1L5Z4 D6WAD3 A0A0T6BEG1 B0X635 A0A1B0FQ23 A0A1A9XB17 A0A067QFH8 A0A1W4X7D9 A0A0M5J110 A0A1Q3FGA9 A0A2J7QRD4 A0A336K3T5 Q16UP8 B8RJG5 A0A1S4FNF8 A0A2P8Y4A2 B3NEV3 A0A182SXB7 A0A182H9F5 A0A182Y1E4 A0A182WID6 A0A182MMT0 Q7Q516 A0A182I4R1 A0A2M4BWS9 A0A1A9URS2 A0A2M4AUG3 A0A2M4BXR0 A0A1W4W4Z3 Q9W086 A0A2M4AUM6 A0A182X4R2 A0A182TPP0 A0A182KAK2 A0A182V443 A0A182PHS3 W5JCA6 A0A1L8DRU2 B4IYN6 A0A182Q6Y9 A0A226EGL8 A0A3B0KAT9 A0A1A9W3E0 B4H5R4 A0A0L0CLN4 A0A2J7QRG5 Q29EZ9 B3M7A5 A0A182FFR5 B4PDU0 A0A0Q9WBR0 B4HW02 A0A1I8M749 A0A0J9RM02 A0A1I8P1E0 A0A1Q3EV02 A0A1L8DRK1 A0A1B6G310 A0A0C9RD06 W8BFV2 A0A0P4X038 A0A1B6IHH3 A0A034WLH0 B4MXT7 A0A1B6KQT0 A0A2S2NTM4 A0A0K8WD24 J3JW99 A0A182JKS2 A0A2R7WAQ8 A0A2H8TKL3 J9JSX7 C4WUX1 A0A182R2F0 A0A0K8TRT2 B4KXU8 U5ES73 A0A1B6DCR1 A0A2P6L0X6 A0A1B0DMP9 A0A0P5YU76 A0A1S3D2Z2 A0A0P6I8C9 A0A2S2QRB6 A0A1B6CUS7 A0A0L8HS31 A0A2M3ZDJ6 A0A2M4AW13 T1IXA6 A0A0J7KSV0 E9FYT6 B4QMK7
A0A212FL90 A0A194R061 A0A1Y1L5Z4 D6WAD3 A0A0T6BEG1 B0X635 A0A1B0FQ23 A0A1A9XB17 A0A067QFH8 A0A1W4X7D9 A0A0M5J110 A0A1Q3FGA9 A0A2J7QRD4 A0A336K3T5 Q16UP8 B8RJG5 A0A1S4FNF8 A0A2P8Y4A2 B3NEV3 A0A182SXB7 A0A182H9F5 A0A182Y1E4 A0A182WID6 A0A182MMT0 Q7Q516 A0A182I4R1 A0A2M4BWS9 A0A1A9URS2 A0A2M4AUG3 A0A2M4BXR0 A0A1W4W4Z3 Q9W086 A0A2M4AUM6 A0A182X4R2 A0A182TPP0 A0A182KAK2 A0A182V443 A0A182PHS3 W5JCA6 A0A1L8DRU2 B4IYN6 A0A182Q6Y9 A0A226EGL8 A0A3B0KAT9 A0A1A9W3E0 B4H5R4 A0A0L0CLN4 A0A2J7QRG5 Q29EZ9 B3M7A5 A0A182FFR5 B4PDU0 A0A0Q9WBR0 B4HW02 A0A1I8M749 A0A0J9RM02 A0A1I8P1E0 A0A1Q3EV02 A0A1L8DRK1 A0A1B6G310 A0A0C9RD06 W8BFV2 A0A0P4X038 A0A1B6IHH3 A0A034WLH0 B4MXT7 A0A1B6KQT0 A0A2S2NTM4 A0A0K8WD24 J3JW99 A0A182JKS2 A0A2R7WAQ8 A0A2H8TKL3 J9JSX7 C4WUX1 A0A182R2F0 A0A0K8TRT2 B4KXU8 U5ES73 A0A1B6DCR1 A0A2P6L0X6 A0A1B0DMP9 A0A0P5YU76 A0A1S3D2Z2 A0A0P6I8C9 A0A2S2QRB6 A0A1B6CUS7 A0A0L8HS31 A0A2M3ZDJ6 A0A2M4AW13 T1IXA6 A0A0J7KSV0 E9FYT6 B4QMK7
Pubmed
19121390
26354079
22651552
22118469
28004739
18362917
+ More
19820115 24845553 17510324 29403074 17994087 26483478 25244985 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 26108605 15632085 17550304 25315136 22936249 24495485 25348373 22516182 23537049 26369729 21292972
19820115 24845553 17510324 29403074 17994087 26483478 25244985 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 26108605 15632085 17550304 25315136 22936249 24495485 25348373 22516182 23537049 26369729 21292972
EMBL
BABH01005696
ODYU01006772
SOQ48928.1
NWSH01000781
PCG74255.1
KQ460927
+ More
KPJ10816.1 AK403211 BAM19634.1 KQ459603 KPI92773.1 AGBW02007871 OWR54460.1 KQ460883 KPJ11188.1 GEZM01066563 JAV67790.1 KQ971312 EEZ98621.2 LJIG01001251 KRT85689.1 DS232400 EDS41166.1 CCAG010009298 KK853582 KDR06498.1 CP012525 ALC44811.1 GFDL01008459 JAV26586.1 NEVH01011898 PNF31154.1 UFQS01000102 UFQT01000102 SSW99644.1 SSX20024.1 CH477616 EAT38260.1 EZ000590 ACJ64464.1 PYGN01000947 PSN39092.1 CH954178 EDV50226.1 JXUM01030164 KQ560874 KXJ80525.1 AXCM01001108 AAAB01008960 EAA11413.3 APCN01002346 GGFJ01008386 MBW57527.1 GGFK01011100 MBW44421.1 GGFJ01008387 MBW57528.1 AE014296 AY071006 AAF47568.1 AAL48628.1 GGFK01011175 MBW44496.1 ADMH02001932 ETN60459.1 GFDF01004987 JAV09097.1 CH916366 EDV95546.1 AXCN02000584 LNIX01000004 OXA56287.1 OUUW01000009 SPP85250.1 CH479211 EDW33116.1 JRES01000206 KNC33278.1 PNF31163.1 CH379070 EAL29910.1 CH902618 EDV38766.1 CM000159 EDW92905.1 CH940682 KRF77568.1 CH480817 EDW50117.1 CM002912 KMY96948.1 GFDL01015903 JAV19142.1 GFDF01004988 JAV09096.1 GECZ01013076 JAS56693.1 GBYB01006175 JAG75942.1 GAMC01014401 GAMC01014399 JAB92156.1 GDRN01040718 JAI67140.1 GECU01021335 JAS86371.1 GAKP01002536 JAC56416.1 CH963876 EDW76856.1 GEBQ01026180 JAT13797.1 GGMR01007886 MBY20505.1 GDHF01008779 GDHF01003253 JAI43535.1 JAI49061.1 APGK01043767 BT127517 KB741017 KB631644 AEE62479.1 ENN75252.1 ERL84937.1 KK854491 PTY16311.1 GFXV01002870 MBW14675.1 ABLF02031057 AK341259 BAH71691.1 GDAI01000531 JAI17072.1 CH933809 EDW17620.1 GANO01003379 JAB56492.1 GEDC01013840 JAS23458.1 MWRG01002814 PRD32222.1 AJVK01007068 AJVK01007069 GDIP01054620 LRGB01000930 JAM49095.1 KZS14958.1 GDIQ01008083 JAN86654.1 GGMS01011078 MBY80281.1 GEDC01020213 JAS17085.1 KQ417510 KOF91555.1 GGFM01005831 MBW26582.1 GGFK01011487 MBW44808.1 JH431646 LBMM01003626 KMQ93309.1 GL732527 EFX87716.1 CM000363 EDX08864.1
KPJ10816.1 AK403211 BAM19634.1 KQ459603 KPI92773.1 AGBW02007871 OWR54460.1 KQ460883 KPJ11188.1 GEZM01066563 JAV67790.1 KQ971312 EEZ98621.2 LJIG01001251 KRT85689.1 DS232400 EDS41166.1 CCAG010009298 KK853582 KDR06498.1 CP012525 ALC44811.1 GFDL01008459 JAV26586.1 NEVH01011898 PNF31154.1 UFQS01000102 UFQT01000102 SSW99644.1 SSX20024.1 CH477616 EAT38260.1 EZ000590 ACJ64464.1 PYGN01000947 PSN39092.1 CH954178 EDV50226.1 JXUM01030164 KQ560874 KXJ80525.1 AXCM01001108 AAAB01008960 EAA11413.3 APCN01002346 GGFJ01008386 MBW57527.1 GGFK01011100 MBW44421.1 GGFJ01008387 MBW57528.1 AE014296 AY071006 AAF47568.1 AAL48628.1 GGFK01011175 MBW44496.1 ADMH02001932 ETN60459.1 GFDF01004987 JAV09097.1 CH916366 EDV95546.1 AXCN02000584 LNIX01000004 OXA56287.1 OUUW01000009 SPP85250.1 CH479211 EDW33116.1 JRES01000206 KNC33278.1 PNF31163.1 CH379070 EAL29910.1 CH902618 EDV38766.1 CM000159 EDW92905.1 CH940682 KRF77568.1 CH480817 EDW50117.1 CM002912 KMY96948.1 GFDL01015903 JAV19142.1 GFDF01004988 JAV09096.1 GECZ01013076 JAS56693.1 GBYB01006175 JAG75942.1 GAMC01014401 GAMC01014399 JAB92156.1 GDRN01040718 JAI67140.1 GECU01021335 JAS86371.1 GAKP01002536 JAC56416.1 CH963876 EDW76856.1 GEBQ01026180 JAT13797.1 GGMR01007886 MBY20505.1 GDHF01008779 GDHF01003253 JAI43535.1 JAI49061.1 APGK01043767 BT127517 KB741017 KB631644 AEE62479.1 ENN75252.1 ERL84937.1 KK854491 PTY16311.1 GFXV01002870 MBW14675.1 ABLF02031057 AK341259 BAH71691.1 GDAI01000531 JAI17072.1 CH933809 EDW17620.1 GANO01003379 JAB56492.1 GEDC01013840 JAS23458.1 MWRG01002814 PRD32222.1 AJVK01007068 AJVK01007069 GDIP01054620 LRGB01000930 JAM49095.1 KZS14958.1 GDIQ01008083 JAN86654.1 GGMS01011078 MBY80281.1 GEDC01020213 JAS17085.1 KQ417510 KOF91555.1 GGFM01005831 MBW26582.1 GGFK01011487 MBW44808.1 JH431646 LBMM01003626 KMQ93309.1 GL732527 EFX87716.1 CM000363 EDX08864.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000007266
+ More
UP000002320 UP000092444 UP000092443 UP000027135 UP000192223 UP000092553 UP000235965 UP000008820 UP000245037 UP000008711 UP000075901 UP000069940 UP000249989 UP000076408 UP000075920 UP000075883 UP000007062 UP000075840 UP000078200 UP000192221 UP000000803 UP000076407 UP000075902 UP000075881 UP000075903 UP000075885 UP000000673 UP000001070 UP000075886 UP000198287 UP000268350 UP000091820 UP000008744 UP000037069 UP000001819 UP000007801 UP000069272 UP000002282 UP000008792 UP000001292 UP000095301 UP000095300 UP000007798 UP000019118 UP000030742 UP000075880 UP000007819 UP000075900 UP000009192 UP000092462 UP000076858 UP000079169 UP000053454 UP000036403 UP000000305 UP000000304
UP000002320 UP000092444 UP000092443 UP000027135 UP000192223 UP000092553 UP000235965 UP000008820 UP000245037 UP000008711 UP000075901 UP000069940 UP000249989 UP000076408 UP000075920 UP000075883 UP000007062 UP000075840 UP000078200 UP000192221 UP000000803 UP000076407 UP000075902 UP000075881 UP000075903 UP000075885 UP000000673 UP000001070 UP000075886 UP000198287 UP000268350 UP000091820 UP000008744 UP000037069 UP000001819 UP000007801 UP000069272 UP000002282 UP000008792 UP000001292 UP000095301 UP000095300 UP000007798 UP000019118 UP000030742 UP000075880 UP000007819 UP000075900 UP000009192 UP000092462 UP000076858 UP000079169 UP000053454 UP000036403 UP000000305 UP000000304
Pfam
Interpro
IPR000086
NUDIX_hydrolase_dom
+ More
IPR040008 MRPL46
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR021757 Ribosomal_L46_N
IPR033650 MRP_L46_NUDIX
IPR016159 Cullin_repeat-like_dom_sf
IPR004140 Exo70
IPR036249 Thioredoxin-like_sf
IPR034732 EPHD
IPR010400 PITH_dom
IPR017937 Thioredoxin_CS
IPR013766 Thioredoxin_domain
IPR037047 PITH_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR008979 Galactose-bd-like_sf
IPR007708 DBR1_C
IPR041677 DNA2/NAM7_AAA_11
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR036058 Kazal_dom_sf
IPR014001 Helicase_ATP-bd
IPR041816 Dbr1_N
IPR002350 Kazal_dom
IPR041679 DNA2/NAM7-like_AAA
IPR007855 RNA-dep_RNA_pol_euk-typ
IPR040008 MRPL46
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR021757 Ribosomal_L46_N
IPR033650 MRP_L46_NUDIX
IPR016159 Cullin_repeat-like_dom_sf
IPR004140 Exo70
IPR036249 Thioredoxin-like_sf
IPR034732 EPHD
IPR010400 PITH_dom
IPR017937 Thioredoxin_CS
IPR013766 Thioredoxin_domain
IPR037047 PITH_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR008979 Galactose-bd-like_sf
IPR007708 DBR1_C
IPR041677 DNA2/NAM7_AAA_11
IPR027417 P-loop_NTPase
IPR003593 AAA+_ATPase
IPR036058 Kazal_dom_sf
IPR014001 Helicase_ATP-bd
IPR041816 Dbr1_N
IPR002350 Kazal_dom
IPR041679 DNA2/NAM7-like_AAA
IPR007855 RNA-dep_RNA_pol_euk-typ
SUPFAM
Gene 3D
ProteinModelPortal
H9JM17
A0A2H1W792
A0A2A4JQB3
A0A0N1I7W3
I4DNZ4
A0A194PIA1
+ More
A0A212FL90 A0A194R061 A0A1Y1L5Z4 D6WAD3 A0A0T6BEG1 B0X635 A0A1B0FQ23 A0A1A9XB17 A0A067QFH8 A0A1W4X7D9 A0A0M5J110 A0A1Q3FGA9 A0A2J7QRD4 A0A336K3T5 Q16UP8 B8RJG5 A0A1S4FNF8 A0A2P8Y4A2 B3NEV3 A0A182SXB7 A0A182H9F5 A0A182Y1E4 A0A182WID6 A0A182MMT0 Q7Q516 A0A182I4R1 A0A2M4BWS9 A0A1A9URS2 A0A2M4AUG3 A0A2M4BXR0 A0A1W4W4Z3 Q9W086 A0A2M4AUM6 A0A182X4R2 A0A182TPP0 A0A182KAK2 A0A182V443 A0A182PHS3 W5JCA6 A0A1L8DRU2 B4IYN6 A0A182Q6Y9 A0A226EGL8 A0A3B0KAT9 A0A1A9W3E0 B4H5R4 A0A0L0CLN4 A0A2J7QRG5 Q29EZ9 B3M7A5 A0A182FFR5 B4PDU0 A0A0Q9WBR0 B4HW02 A0A1I8M749 A0A0J9RM02 A0A1I8P1E0 A0A1Q3EV02 A0A1L8DRK1 A0A1B6G310 A0A0C9RD06 W8BFV2 A0A0P4X038 A0A1B6IHH3 A0A034WLH0 B4MXT7 A0A1B6KQT0 A0A2S2NTM4 A0A0K8WD24 J3JW99 A0A182JKS2 A0A2R7WAQ8 A0A2H8TKL3 J9JSX7 C4WUX1 A0A182R2F0 A0A0K8TRT2 B4KXU8 U5ES73 A0A1B6DCR1 A0A2P6L0X6 A0A1B0DMP9 A0A0P5YU76 A0A1S3D2Z2 A0A0P6I8C9 A0A2S2QRB6 A0A1B6CUS7 A0A0L8HS31 A0A2M3ZDJ6 A0A2M4AW13 T1IXA6 A0A0J7KSV0 E9FYT6 B4QMK7
A0A212FL90 A0A194R061 A0A1Y1L5Z4 D6WAD3 A0A0T6BEG1 B0X635 A0A1B0FQ23 A0A1A9XB17 A0A067QFH8 A0A1W4X7D9 A0A0M5J110 A0A1Q3FGA9 A0A2J7QRD4 A0A336K3T5 Q16UP8 B8RJG5 A0A1S4FNF8 A0A2P8Y4A2 B3NEV3 A0A182SXB7 A0A182H9F5 A0A182Y1E4 A0A182WID6 A0A182MMT0 Q7Q516 A0A182I4R1 A0A2M4BWS9 A0A1A9URS2 A0A2M4AUG3 A0A2M4BXR0 A0A1W4W4Z3 Q9W086 A0A2M4AUM6 A0A182X4R2 A0A182TPP0 A0A182KAK2 A0A182V443 A0A182PHS3 W5JCA6 A0A1L8DRU2 B4IYN6 A0A182Q6Y9 A0A226EGL8 A0A3B0KAT9 A0A1A9W3E0 B4H5R4 A0A0L0CLN4 A0A2J7QRG5 Q29EZ9 B3M7A5 A0A182FFR5 B4PDU0 A0A0Q9WBR0 B4HW02 A0A1I8M749 A0A0J9RM02 A0A1I8P1E0 A0A1Q3EV02 A0A1L8DRK1 A0A1B6G310 A0A0C9RD06 W8BFV2 A0A0P4X038 A0A1B6IHH3 A0A034WLH0 B4MXT7 A0A1B6KQT0 A0A2S2NTM4 A0A0K8WD24 J3JW99 A0A182JKS2 A0A2R7WAQ8 A0A2H8TKL3 J9JSX7 C4WUX1 A0A182R2F0 A0A0K8TRT2 B4KXU8 U5ES73 A0A1B6DCR1 A0A2P6L0X6 A0A1B0DMP9 A0A0P5YU76 A0A1S3D2Z2 A0A0P6I8C9 A0A2S2QRB6 A0A1B6CUS7 A0A0L8HS31 A0A2M3ZDJ6 A0A2M4AW13 T1IXA6 A0A0J7KSV0 E9FYT6 B4QMK7
PDB
6GB2
E-value=1.03035e-41,
Score=425
Ontologies
GO
Topology
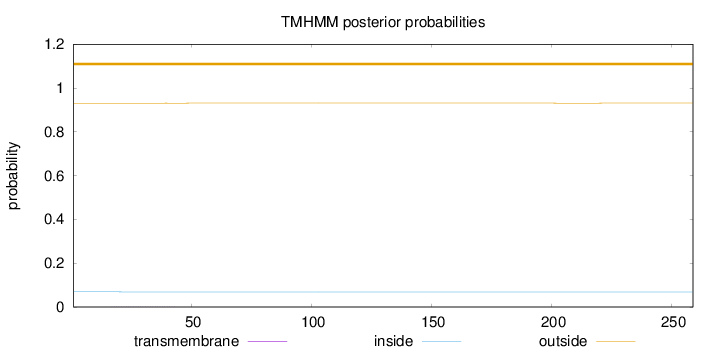
Length:
259
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00882
Exp number, first 60 AAs:
0.00825
Total prob of N-in:
0.06924
outside
1 - 259
Population Genetic Test Statistics
Pi
74.827587
Theta
128.318513
Tajima's D
-0.668566
CLR
0.620663
CSRT
0.204139793010349
Interpretation
Uncertain
