Pre Gene Modal
BGIBMGA010385
Annotation
PREDICTED:_non-lysosomal_glucosylceramidase-like_isoform_X2_[Bombyx_mori]
Full name
Non-lysosomal glucosylceramidase
Location in the cell
Mitochondrial Reliability : 2.108
Sequence
CDS
ATGGATATCGAACCCGCAGCCAAAGAACACCGCGATTTTGAAATGGACAAACCATCGAAATTCGGCTTAAAACTTCCCTTAGATCATGAATTTCCCCTGAAGAGCAAGCAGATTATAATTCCAAGACCAAAACAAATACTGGACATGATACCGCTTTCAGTCAGGTATTTCAAGTATTGTTTGAGCAAAAAACTGCGTAAAAGGCGTCCAATAATGGATTATTTCCAAATGATTTCAGCACAGCGAATGTACGGATGTCCCATTGGTGGAATTGGAGGTGGAACCATAGGGAGGGGTTTTAAAGGGGAATTTTGTCGTTTTCAACTACATCCTGGTATATATGAATATGTCACTGTCCCAGAATGCCAGTTTATTGTAAACATAAGGAATGCTAACAATGAGACAATATTCCAATCAGTTCTTTCCACCTACAGCAAACCAAAGAAAGCCCCGGCATCTTGGGAATGGAACCTTGATGGTGCAAAATGTGAATACACAGCACTGTATCCGAGGGCATGGACAACATATGATCTGTCAAAATATGGTGTGAAACTTGTGTGTAGACAAATATCACCAGTAATACCTCATAACTATAAGGACAGCAAGACGTGA
Protein
MDIEPAAKEHRDFEMDKPSKFGLKLPLDHEFPLKSKQIIIPRPKQILDMIPLSVRYFKYCLSKKLRKRRPIMDYFQMISAQRMYGCPIGGIGGGTIGRGFKGEFCRFQLHPGIYEYVTVPECQFIVNIRNANNETIFQSVLSTYSKPKKAPASWEWNLDGAKCEYTALYPRAWTTYDLSKYGVKLVCRQISPVIPHNYKDSKT
Summary
Description
Non-lysosomal glucosylceramidase that catalyzes the hydrolysis of glucosylceramide (GlcCer) to free glucose and ceramide.
Non-lysosomal glucosylceramidase that catalyzes the conversion of glucosylceramide to free glucose and ceramide.
Non-lysosomal glucosylceramidase that catalyzes the conversion of glucosylceramide to free glucose and ceramide.
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the non-lysosomal glucosylceramidase family.
Keywords
Cell membrane
Complete proteome
Glycoprotein
Glycosidase
Hydrolase
Lipid metabolism
Membrane
Phosphoprotein
Reference proteome
Sphingolipid metabolism
Transmembrane
Transmembrane helix
Feature
chain Non-lysosomal glucosylceramidase
Uniprot
H9JLI4
A0A2A4JS75
A0A194PHH7
A0A194R067
A0A3S2MAG2
A0A212FL48
+ More
A0A1J1J8E5 A0A1Q3EX63 A0A2M4AD48 A0A182G2J8 D6WP98 A0A182VG64 A0A182XDH2 A0A1B0D2U1 A0A182F4Y7 A0A2M4CTR6 A0A1B0CJL7 A0A1Y1M7I9 A0A1S4EYW6 A0A1Y1M6Q4 A0A336LV92 A0A0N7ZAY4 A0A182J903 A0A1I8Q3I3 A0A1L8E2K8 A0A1L8E229 A0A232FNF0 A0A182Y986 A0A336KNI8 A0A2S2NZ18 A0A1I8MB60 A0A2S2NJ98 A0A1A9X643 K7J4S3 A0A0A1X3L1 A0A0Q9W6A9 W8BBN7 A0A1I8Q3F9 A0A0L0BZ66 A0A084VYE4 A0A2C9GPH9 A0A1Y1M358 A0A1S4GZD2 A0A2H8TUQ7 A0A1I8MB61 A0A1B0BIX2 A0A0A1WH86 A0A0Q9XEM2 A0A1W4W6S7 A0A0K8VHU5 A0A0R1DLK3 A0A1B0AJL2 A0A034VAV9 A0A182Q9Z3 T1JDY2 Q7PWW0 A0A0K8U0T0 A0A1W4UXX5 A0A034VCF4 Q17L47 A0A0R3NV91 A0A3B0JQ10 W8BYU9 A0A0Q5WAH1 X1WIY6 B0WC06 E0VQ36 A0A0P8Y3U4 N6TMK2 X2JAF7 A0A0J9TN83 A0A182VU42 A0A2S2QCY2 B4MZB3 A0A1A9VQD2 A0A1B6DQC9 B4P0J9 A0A1B6DSU6 A0A1B6CFK6 A0A1Y1M3J2 A0A1B6GW49 A0A1W4UJ88 B5DJY3 A0A182RUT0 A0A0Q5WJC6 A0A3B0JNL6 B3N9T4 X2J9U5 A0A0J9R344 Q7KT91 B4Q5I4 B4HXF9 A0A0J9R2C1 A0A182MMM5 B3MKK0 A0A2T7NF00 A0A1B6LPM5
A0A1J1J8E5 A0A1Q3EX63 A0A2M4AD48 A0A182G2J8 D6WP98 A0A182VG64 A0A182XDH2 A0A1B0D2U1 A0A182F4Y7 A0A2M4CTR6 A0A1B0CJL7 A0A1Y1M7I9 A0A1S4EYW6 A0A1Y1M6Q4 A0A336LV92 A0A0N7ZAY4 A0A182J903 A0A1I8Q3I3 A0A1L8E2K8 A0A1L8E229 A0A232FNF0 A0A182Y986 A0A336KNI8 A0A2S2NZ18 A0A1I8MB60 A0A2S2NJ98 A0A1A9X643 K7J4S3 A0A0A1X3L1 A0A0Q9W6A9 W8BBN7 A0A1I8Q3F9 A0A0L0BZ66 A0A084VYE4 A0A2C9GPH9 A0A1Y1M358 A0A1S4GZD2 A0A2H8TUQ7 A0A1I8MB61 A0A1B0BIX2 A0A0A1WH86 A0A0Q9XEM2 A0A1W4W6S7 A0A0K8VHU5 A0A0R1DLK3 A0A1B0AJL2 A0A034VAV9 A0A182Q9Z3 T1JDY2 Q7PWW0 A0A0K8U0T0 A0A1W4UXX5 A0A034VCF4 Q17L47 A0A0R3NV91 A0A3B0JQ10 W8BYU9 A0A0Q5WAH1 X1WIY6 B0WC06 E0VQ36 A0A0P8Y3U4 N6TMK2 X2JAF7 A0A0J9TN83 A0A182VU42 A0A2S2QCY2 B4MZB3 A0A1A9VQD2 A0A1B6DQC9 B4P0J9 A0A1B6DSU6 A0A1B6CFK6 A0A1Y1M3J2 A0A1B6GW49 A0A1W4UJ88 B5DJY3 A0A182RUT0 A0A0Q5WJC6 A0A3B0JNL6 B3N9T4 X2J9U5 A0A0J9R344 Q7KT91 B4Q5I4 B4HXF9 A0A0J9R2C1 A0A182MMM5 B3MKK0 A0A2T7NF00 A0A1B6LPM5
EC Number
3.2.1.45
Pubmed
19121390
26354079
22118469
26483478
18362917
19820115
+ More
28004739 17510324 28648823 25244985 25315136 20075255 25830018 17994087 24495485 26108605 24438588 12364791 17550304 25348373 15632085 20566863 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 12537569 18327897
28004739 17510324 28648823 25244985 25315136 20075255 25830018 17994087 24495485 26108605 24438588 12364791 17550304 25348373 15632085 20566863 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 12537569 18327897
EMBL
BABH01005693
BABH01005694
BABH01005695
BABH01005696
NWSH01000781
PCG74252.1
+ More
KQ459603 KPI92767.1 KQ460883 KPJ11193.1 RSAL01000003 RVE54749.1 AGBW02007871 OWR54456.1 CVRI01000074 CRL08234.1 GFDL01015179 JAV19866.1 GGFK01005378 MBW38699.1 JXUM01040103 JXUM01040104 JXUM01040105 JXUM01040106 KQ561232 KXJ79285.1 KQ971354 EFA07388.1 AJVK01002842 AJVK01002843 AJVK01002844 GGFL01004568 MBW68746.1 AJWK01014724 AJWK01014725 AJWK01014726 AJWK01014727 GEZM01041578 JAV80275.1 GEZM01041586 JAV80270.1 UFQT01000222 SSX21962.1 GDRN01092272 GDRN01092271 JAI60136.1 GFDF01001349 JAV12735.1 GFDF01001350 JAV12734.1 NNAY01000010 OXU32049.1 UFQS01000634 UFQT01000634 SSX05588.1 SSX25947.1 GGMR01009796 MBY22415.1 GGMR01004635 MBY17254.1 AAZX01005602 GBXI01008373 JAD05919.1 CH940654 KRF77831.1 GAMC01012127 JAB94428.1 JRES01001125 KNC25310.1 ATLV01018361 ATLV01018362 ATLV01018363 KE525231 KFB42988.1 APCN01003341 APCN01003342 GEZM01041579 JAV80274.1 AAAB01008984 GFXV01005103 MBW16908.1 JXJN01015223 GBXI01016236 JAC98055.1 CH933807 KRG02958.1 GDHF01013840 JAI38474.1 CM000157 KRJ98122.1 GAKP01019748 GAKP01019747 GAKP01019745 GAKP01019744 JAC39207.1 AXCN02000732 JH432114 EAA14848.4 GDHF01031985 JAI20329.1 GAKP01019749 GAKP01019746 JAC39203.1 CH477219 EAT47379.1 CH379061 KRT04860.1 OUUW01000006 SPP82482.1 GAMC01012126 JAB94429.1 CH954177 KQS70445.1 ABLF02018149 ABLF02018157 ABLF02018163 ABLF02059614 DS231884 EDS43066.1 DS235389 EEB15492.1 CH902620 KPU73469.1 APGK01029406 APGK01029407 APGK01029408 APGK01029409 KB740734 ENN79258.1 AE014134 AHN54442.1 CM002910 KMY90235.1 GGMS01006401 MBY75604.1 CH963913 EDW77386.1 GEDC01009404 JAS27894.1 EDW88993.1 GEDC01008565 JAS28733.1 GEDC01025021 JAS12277.1 GEZM01041580 JAV80273.1 GECZ01003088 JAS66681.1 EDY70607.2 KQS70446.1 SPP82483.1 EDV58579.1 AHN54441.1 KMY90234.1 BT024218 BT044085 AY058463 CM000361 EDX05028.1 CH480818 EDW51739.1 KMY90233.1 AXCM01000045 EDV31553.2 PZQS01000013 PVD19748.1 GEBQ01014339 GEBQ01001449 JAT25638.1 JAT38528.1
KQ459603 KPI92767.1 KQ460883 KPJ11193.1 RSAL01000003 RVE54749.1 AGBW02007871 OWR54456.1 CVRI01000074 CRL08234.1 GFDL01015179 JAV19866.1 GGFK01005378 MBW38699.1 JXUM01040103 JXUM01040104 JXUM01040105 JXUM01040106 KQ561232 KXJ79285.1 KQ971354 EFA07388.1 AJVK01002842 AJVK01002843 AJVK01002844 GGFL01004568 MBW68746.1 AJWK01014724 AJWK01014725 AJWK01014726 AJWK01014727 GEZM01041578 JAV80275.1 GEZM01041586 JAV80270.1 UFQT01000222 SSX21962.1 GDRN01092272 GDRN01092271 JAI60136.1 GFDF01001349 JAV12735.1 GFDF01001350 JAV12734.1 NNAY01000010 OXU32049.1 UFQS01000634 UFQT01000634 SSX05588.1 SSX25947.1 GGMR01009796 MBY22415.1 GGMR01004635 MBY17254.1 AAZX01005602 GBXI01008373 JAD05919.1 CH940654 KRF77831.1 GAMC01012127 JAB94428.1 JRES01001125 KNC25310.1 ATLV01018361 ATLV01018362 ATLV01018363 KE525231 KFB42988.1 APCN01003341 APCN01003342 GEZM01041579 JAV80274.1 AAAB01008984 GFXV01005103 MBW16908.1 JXJN01015223 GBXI01016236 JAC98055.1 CH933807 KRG02958.1 GDHF01013840 JAI38474.1 CM000157 KRJ98122.1 GAKP01019748 GAKP01019747 GAKP01019745 GAKP01019744 JAC39207.1 AXCN02000732 JH432114 EAA14848.4 GDHF01031985 JAI20329.1 GAKP01019749 GAKP01019746 JAC39203.1 CH477219 EAT47379.1 CH379061 KRT04860.1 OUUW01000006 SPP82482.1 GAMC01012126 JAB94429.1 CH954177 KQS70445.1 ABLF02018149 ABLF02018157 ABLF02018163 ABLF02059614 DS231884 EDS43066.1 DS235389 EEB15492.1 CH902620 KPU73469.1 APGK01029406 APGK01029407 APGK01029408 APGK01029409 KB740734 ENN79258.1 AE014134 AHN54442.1 CM002910 KMY90235.1 GGMS01006401 MBY75604.1 CH963913 EDW77386.1 GEDC01009404 JAS27894.1 EDW88993.1 GEDC01008565 JAS28733.1 GEDC01025021 JAS12277.1 GEZM01041580 JAV80273.1 GECZ01003088 JAS66681.1 EDY70607.2 KQS70446.1 SPP82483.1 EDV58579.1 AHN54441.1 KMY90234.1 BT024218 BT044085 AY058463 CM000361 EDX05028.1 CH480818 EDW51739.1 KMY90233.1 AXCM01000045 EDV31553.2 PZQS01000013 PVD19748.1 GEBQ01014339 GEBQ01001449 JAT25638.1 JAT38528.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000183832 UP000069940 UP000249989 UP000007266 UP000075903 UP000076407 UP000092462 UP000069272 UP000092461 UP000075880 UP000095300 UP000215335 UP000076408 UP000095301 UP000092443 UP000002358 UP000008792 UP000037069 UP000030765 UP000075840 UP000092460 UP000009192 UP000192223 UP000002282 UP000092445 UP000075886 UP000007062 UP000192221 UP000008820 UP000001819 UP000268350 UP000008711 UP000007819 UP000002320 UP000009046 UP000007801 UP000019118 UP000000803 UP000075920 UP000007798 UP000078200 UP000075900 UP000000304 UP000001292 UP000075883 UP000245119
UP000183832 UP000069940 UP000249989 UP000007266 UP000075903 UP000076407 UP000092462 UP000069272 UP000092461 UP000075880 UP000095300 UP000215335 UP000076408 UP000095301 UP000092443 UP000002358 UP000008792 UP000037069 UP000030765 UP000075840 UP000092460 UP000009192 UP000192223 UP000002282 UP000092445 UP000075886 UP000007062 UP000192221 UP000008820 UP000001819 UP000268350 UP000008711 UP000007819 UP000002320 UP000009046 UP000007801 UP000019118 UP000000803 UP000075920 UP000007798 UP000078200 UP000075900 UP000000304 UP000001292 UP000075883 UP000245119
Pfam
Interpro
IPR024462
GH116_N
+ More
IPR012341 6hp_glycosidase-like_sf
IPR008928 6-hairpin_glycosidase_sf
IPR006775 GH116_catalytic
IPR014551 B_Glucosidase_GBA2-typ
IPR003617 TFIIS/CRSP70_N_sub
IPR006289 TFSII
IPR031567 CRIM_dom
IPR032679 Sin1_N
IPR017923 TFIIS_N
IPR001222 Znf_TFIIS
IPR003618 TFIIS_cen_dom
IPR011993 PH-like_dom_sf
IPR036575 TFIIS_cen_dom_sf
IPR035441 TFIIS/LEDGF_dom_sf
IPR031313 Sin1_PH_dom
IPR019591 Mrp/NBP35_ATP-bd
IPR027417 P-loop_NTPase
IPR033756 YlxH/NBP35
IPR000808 Mrp_CS
IPR012341 6hp_glycosidase-like_sf
IPR008928 6-hairpin_glycosidase_sf
IPR006775 GH116_catalytic
IPR014551 B_Glucosidase_GBA2-typ
IPR003617 TFIIS/CRSP70_N_sub
IPR006289 TFSII
IPR031567 CRIM_dom
IPR032679 Sin1_N
IPR017923 TFIIS_N
IPR001222 Znf_TFIIS
IPR003618 TFIIS_cen_dom
IPR011993 PH-like_dom_sf
IPR036575 TFIIS_cen_dom_sf
IPR035441 TFIIS/LEDGF_dom_sf
IPR031313 Sin1_PH_dom
IPR019591 Mrp/NBP35_ATP-bd
IPR027417 P-loop_NTPase
IPR033756 YlxH/NBP35
IPR000808 Mrp_CS
Gene 3D
CDD
ProteinModelPortal
H9JLI4
A0A2A4JS75
A0A194PHH7
A0A194R067
A0A3S2MAG2
A0A212FL48
+ More
A0A1J1J8E5 A0A1Q3EX63 A0A2M4AD48 A0A182G2J8 D6WP98 A0A182VG64 A0A182XDH2 A0A1B0D2U1 A0A182F4Y7 A0A2M4CTR6 A0A1B0CJL7 A0A1Y1M7I9 A0A1S4EYW6 A0A1Y1M6Q4 A0A336LV92 A0A0N7ZAY4 A0A182J903 A0A1I8Q3I3 A0A1L8E2K8 A0A1L8E229 A0A232FNF0 A0A182Y986 A0A336KNI8 A0A2S2NZ18 A0A1I8MB60 A0A2S2NJ98 A0A1A9X643 K7J4S3 A0A0A1X3L1 A0A0Q9W6A9 W8BBN7 A0A1I8Q3F9 A0A0L0BZ66 A0A084VYE4 A0A2C9GPH9 A0A1Y1M358 A0A1S4GZD2 A0A2H8TUQ7 A0A1I8MB61 A0A1B0BIX2 A0A0A1WH86 A0A0Q9XEM2 A0A1W4W6S7 A0A0K8VHU5 A0A0R1DLK3 A0A1B0AJL2 A0A034VAV9 A0A182Q9Z3 T1JDY2 Q7PWW0 A0A0K8U0T0 A0A1W4UXX5 A0A034VCF4 Q17L47 A0A0R3NV91 A0A3B0JQ10 W8BYU9 A0A0Q5WAH1 X1WIY6 B0WC06 E0VQ36 A0A0P8Y3U4 N6TMK2 X2JAF7 A0A0J9TN83 A0A182VU42 A0A2S2QCY2 B4MZB3 A0A1A9VQD2 A0A1B6DQC9 B4P0J9 A0A1B6DSU6 A0A1B6CFK6 A0A1Y1M3J2 A0A1B6GW49 A0A1W4UJ88 B5DJY3 A0A182RUT0 A0A0Q5WJC6 A0A3B0JNL6 B3N9T4 X2J9U5 A0A0J9R344 Q7KT91 B4Q5I4 B4HXF9 A0A0J9R2C1 A0A182MMM5 B3MKK0 A0A2T7NF00 A0A1B6LPM5
A0A1J1J8E5 A0A1Q3EX63 A0A2M4AD48 A0A182G2J8 D6WP98 A0A182VG64 A0A182XDH2 A0A1B0D2U1 A0A182F4Y7 A0A2M4CTR6 A0A1B0CJL7 A0A1Y1M7I9 A0A1S4EYW6 A0A1Y1M6Q4 A0A336LV92 A0A0N7ZAY4 A0A182J903 A0A1I8Q3I3 A0A1L8E2K8 A0A1L8E229 A0A232FNF0 A0A182Y986 A0A336KNI8 A0A2S2NZ18 A0A1I8MB60 A0A2S2NJ98 A0A1A9X643 K7J4S3 A0A0A1X3L1 A0A0Q9W6A9 W8BBN7 A0A1I8Q3F9 A0A0L0BZ66 A0A084VYE4 A0A2C9GPH9 A0A1Y1M358 A0A1S4GZD2 A0A2H8TUQ7 A0A1I8MB61 A0A1B0BIX2 A0A0A1WH86 A0A0Q9XEM2 A0A1W4W6S7 A0A0K8VHU5 A0A0R1DLK3 A0A1B0AJL2 A0A034VAV9 A0A182Q9Z3 T1JDY2 Q7PWW0 A0A0K8U0T0 A0A1W4UXX5 A0A034VCF4 Q17L47 A0A0R3NV91 A0A3B0JQ10 W8BYU9 A0A0Q5WAH1 X1WIY6 B0WC06 E0VQ36 A0A0P8Y3U4 N6TMK2 X2JAF7 A0A0J9TN83 A0A182VU42 A0A2S2QCY2 B4MZB3 A0A1A9VQD2 A0A1B6DQC9 B4P0J9 A0A1B6DSU6 A0A1B6CFK6 A0A1Y1M3J2 A0A1B6GW49 A0A1W4UJ88 B5DJY3 A0A182RUT0 A0A0Q5WJC6 A0A3B0JNL6 B3N9T4 X2J9U5 A0A0J9R344 Q7KT91 B4Q5I4 B4HXF9 A0A0J9R2C1 A0A182MMM5 B3MKK0 A0A2T7NF00 A0A1B6LPM5
PDB
5OST
E-value=8.69048e-10,
Score=149
Ontologies
PATHWAY
GO
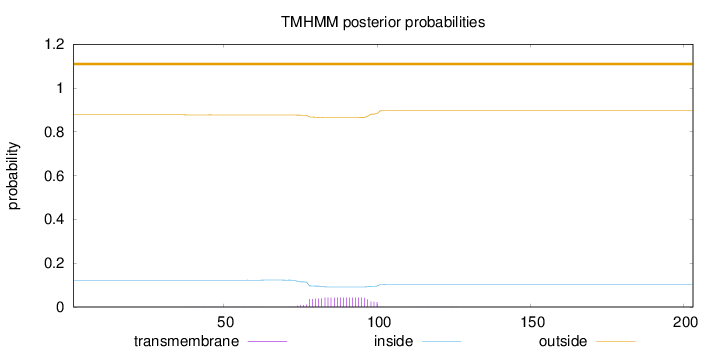
Topology
Subcellular location
Nucleus
Cell membrane
Cell membrane
Length:
203
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.951
Exp number, first 60 AAs:
0.01038
Total prob of N-in:
0.12224
outside
1 - 203
Population Genetic Test Statistics
Pi
15.774678
Theta
14.593288
Tajima's D
-0.911977
CLR
3.114168
CSRT
0.156442177891105
Interpretation
Uncertain
