Pre Gene Modal
BGIBMGA010385
Annotation
PREDICTED:_non-lysosomal_glucosylceramidase-like_isoform_X2_[Bombyx_mori]
Full name
Non-lysosomal glucosylceramidase
Location in the cell
Cytoplasmic Reliability : 2.187 Nuclear Reliability : 1.609
Sequence
CDS
ATGAATAAAATCGATTTGGAAAGGTATTCTGAGGACTATACGTGCGGAGTTTCGTTAGAGCAAAGAATCGGCGAAAGCTTATGTACGTTTACGATAGCGAAAAAGAAAGTCGATGACGAAACGATCCACCAGGGCTATTGCCTGTGGAACGCGGCGAAGACTTCCAACTACGTTTGGGAATGCATAAAGAAAGATGGGAAGCTAACTCCGGATCCAAACTTAACTCCACCGCCGCCCAAACTTCCCGACAAAGACGCACAAAAGAAAAAGAAAGACAAAGAAAAAAAGAAACTGTATAAATGTGATTCAGTCGCCCTGTCCAGCGTCATATCGTTAGATCCAGGGGGAGTTAGTTCAACGGAGTTTTGTCTGGCCTGGGATATGCCTGTAGTTAAATATAGAAACGACAAAAAAATTCACAGAAGATTTTACACGAAATATTTTGGTAGTGACGGCATAGCCGGTGCGAGCATAGCTGCCTACGCTCTAAAGCATTACAAAAATTGGGAAAAACAGCTTCTTGAATGGCAGGAGCCAATTTTAACTAATTACAATATACCAGACTGGTTGAAAAGTGCCTTAATGAACGAGCTTTATTTTGTCGCCGATGGCGGAACGATTTGGTTTGATGCAAGCGACGAATTTCCAGAAACGGATCCTAGGTAA
Protein
MNKIDLERYSEDYTCGVSLEQRIGESLCTFTIAKKKVDDETIHQGYCLWNAAKTSNYVWECIKKDGKLTPDPNLTPPPPKLPDKDAQKKKKDKEKKKLYKCDSVALSSVISLDPGGVSSTEFCLAWDMPVVKYRNDKKIHRRFYTKYFGSDGIAGASIAAYALKHYKNWEKQLLEWQEPILTNYNIPDWLKSALMNELYFVADGGTIWFDASDEFPETDPR
Summary
Description
Non-lysosomal glucosylceramidase that catalyzes the hydrolysis of glucosylceramide (GlcCer) to free glucose and ceramide.
Catalytic Activity
a beta-D-glucosyl-(1<->1')-N-acylsphing-4-enine + H2O = an N-acylsphing-4-enine + D-glucose
Similarity
Belongs to the non-lysosomal glucosylceramidase family.
Uniprot
A0A194R067
A0A194PHH7
A0A2H1WWR2
A0A212EKV0
A0A212FL48
A0A3S2MAG2
+ More
A0A1E1WJ95 A0A0L7KYM6 A0A1L8E2K8 A0A2C9GPH9 A0A1L8E229 A0A1I8MB61 A0A1I8MB60 W5JIF6 A0A2M4CTR6 A0A0L0BZ66 A0A2M4AD48 A0A1Y1M339 A0A1I8Q3F9 A0A1Y1M8M0 A0A1Y1M460 A0A1I8Q3I3 A0A1Y1M7I9 B4MZB3 A0A1Y1M6Q4 A0A1Y1M3J2 A0A1Y1M358 B4P0J9
A0A1E1WJ95 A0A0L7KYM6 A0A1L8E2K8 A0A2C9GPH9 A0A1L8E229 A0A1I8MB61 A0A1I8MB60 W5JIF6 A0A2M4CTR6 A0A0L0BZ66 A0A2M4AD48 A0A1Y1M339 A0A1I8Q3F9 A0A1Y1M8M0 A0A1Y1M460 A0A1I8Q3I3 A0A1Y1M7I9 B4MZB3 A0A1Y1M6Q4 A0A1Y1M3J2 A0A1Y1M358 B4P0J9
EC Number
3.2.1.45
Pubmed
EMBL
KQ460883
KPJ11193.1
KQ459603
KPI92767.1
ODYU01011634
SOQ57503.1
+ More
AGBW02014207 OWR42113.1 AGBW02007871 OWR54456.1 RSAL01000003 RVE54749.1 GDQN01003974 JAT87080.1 JTDY01004287 KOB68332.1 GFDF01001349 JAV12735.1 APCN01003341 APCN01003342 GFDF01001350 JAV12734.1 ADMH02001302 ETN63073.1 GGFL01004568 MBW68746.1 JRES01001125 KNC25310.1 GGFK01005378 MBW38699.1 GEZM01041582 JAV80272.1 GEZM01041577 JAV80276.1 GEZM01041585 JAV80271.1 GEZM01041578 JAV80275.1 CH963913 EDW77386.1 GEZM01041586 JAV80270.1 GEZM01041580 JAV80273.1 GEZM01041579 JAV80274.1 CM000157 EDW88993.1
AGBW02014207 OWR42113.1 AGBW02007871 OWR54456.1 RSAL01000003 RVE54749.1 GDQN01003974 JAT87080.1 JTDY01004287 KOB68332.1 GFDF01001349 JAV12735.1 APCN01003341 APCN01003342 GFDF01001350 JAV12734.1 ADMH02001302 ETN63073.1 GGFL01004568 MBW68746.1 JRES01001125 KNC25310.1 GGFK01005378 MBW38699.1 GEZM01041582 JAV80272.1 GEZM01041577 JAV80276.1 GEZM01041585 JAV80271.1 GEZM01041578 JAV80275.1 CH963913 EDW77386.1 GEZM01041586 JAV80270.1 GEZM01041580 JAV80273.1 GEZM01041579 JAV80274.1 CM000157 EDW88993.1
Proteomes
Interpro
SUPFAM
SSF48208
SSF48208
Gene 3D
ProteinModelPortal
A0A194R067
A0A194PHH7
A0A2H1WWR2
A0A212EKV0
A0A212FL48
A0A3S2MAG2
+ More
A0A1E1WJ95 A0A0L7KYM6 A0A1L8E2K8 A0A2C9GPH9 A0A1L8E229 A0A1I8MB61 A0A1I8MB60 W5JIF6 A0A2M4CTR6 A0A0L0BZ66 A0A2M4AD48 A0A1Y1M339 A0A1I8Q3F9 A0A1Y1M8M0 A0A1Y1M460 A0A1I8Q3I3 A0A1Y1M7I9 B4MZB3 A0A1Y1M6Q4 A0A1Y1M3J2 A0A1Y1M358 B4P0J9
A0A1E1WJ95 A0A0L7KYM6 A0A1L8E2K8 A0A2C9GPH9 A0A1L8E229 A0A1I8MB61 A0A1I8MB60 W5JIF6 A0A2M4CTR6 A0A0L0BZ66 A0A2M4AD48 A0A1Y1M339 A0A1I8Q3F9 A0A1Y1M8M0 A0A1Y1M460 A0A1I8Q3I3 A0A1Y1M7I9 B4MZB3 A0A1Y1M6Q4 A0A1Y1M3J2 A0A1Y1M358 B4P0J9
PDB
5OST
E-value=1.71818e-18,
Score=224
Ontologies
Topology
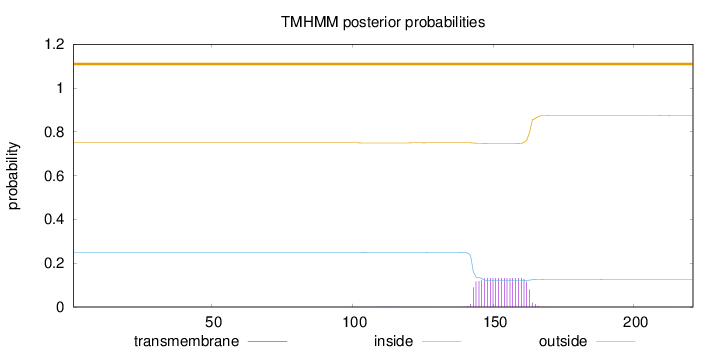
Length:
221
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.71859
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.25037
outside
1 - 221
Population Genetic Test Statistics
Pi
50.045779
Theta
83.931124
Tajima's D
-1.170216
CLR
76.870641
CSRT
0.107944602769862
Interpretation
Uncertain
