Gene
KWMTBOMO07190 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010387
Annotation
DnaJ-12_[Bombyx_mori]
Full name
J domain-containing protein
Alternative Name
dJDP
Location in the cell
Nuclear Reliability : 3.715
Sequence
CDS
ATGACTGGCGTAGACGAAATATTGAATTATCAAAGAAACCCAGATGATGACTACTACGCCCTACTGGGTTGTGATGAAAACTCAACTGTGGAGCAAATAACTGCAGAATATAAGATACTTGCCCTGCAACACCACCCTGACAAAAACGATGGTGAAAAAGAAGCTGAAATGAAGTTTCAGAAGTTGAAGGAAGCCAAGGAGATTCTGTGTGATCCATCCAAGCGTGCTCTGTATGACAAGTGGCGTCGTAGTGGTATAGCTATGGGCTTCAAGCAGTGGTTGGGTATGAAAGATCATGTGCAGCAGTCCATGCACTGGTCCAAGCCTAATACTAAGGATCGCATGTTGGAAGGAGATGGAAGCAAGCCGTCAGGTCCATCTAGCCTTGGCCCCAGTAACCCTGCTACACGTCGGGCTTCTGAGGGTGGAGCTGCTCTCTGGGGACGCTGGGGAACTGGCAATCAAGAACCACCCTCTGAGGTCCTCTCCAAGTTTCGCAACTATGAAATCTAA
Protein
MTGVDEILNYQRNPDDDYYALLGCDENSTVEQITAEYKILALQHHPDKNDGEKEAEMKFQKLKEAKEILCDPSKRALYDKWRRSGIAMGFKQWLGMKDHVQQSMHWSKPNTKDRMLEGDGSKPSGPSSLGPSNPATRRASEGGAALWGRWGTGNQEPPSEVLSKFRNYEI
Summary
Keywords
Chaperone
Complete proteome
Reference proteome
Alternative splicing
Feature
chain J domain-containing protein
splice variant In isoform A.
splice variant In isoform A.
Uniprot
C7AR00
Q9U6V7
H6VTQ2
H9JLI6
A0A194R1S0
A0A194PI92
+ More
Q9U6V6 A0A2H1WWQ8 S4PTE0 A0A2A4K1K7 A0A212EKW0 D6X3Z1 U5EUF7 A0A023EIU1 Q173S6 A0A1L8DUB4 A0A1Y1NH90 A0A1Q3G347 A0A1Q3G3A3 A0A1A9ZZK6 D3TRX1 W8BBJ0 A0A1B6MKP8 A0A034V3D6 A0A1I8QAY6 A0A182Q044 A0A0Q9X7A7 T1P818 A0A182W448 A0A1B6DPJ5 Q7QI07 A0A0K8UJK9 A0A0P8XUA6 A0A182RH62 A0A0A1X2A2 A0A154PFV2 A0A182JGC1 E2BFX9 B4HZS7 A0A084VN13 B4PMI8 Q9TVP3-2 H0RNN1 A0A182PPX2 B4R273 A0A182XNH9 A0A182V6C5 A0A182IDP0 A0A224XRB5 A0A069DPM1 B3P570 A0A0Q9WNV7 A0A023F8Q1 N6U8N6 A0A0Q9XAH2 A0A0N7Z9D6 T1HJG3 A0A182U6U9 A0A1W4WP47 A0A1A9VVH1 K7J003 A0A1W4W2P0 T1DS84 A0A182Y543 A0A1B0FAK0 A0A0J7NRY7 A0A2J7RPR6 A0A2M4C0W3 A0A336KZ16 A0A182FIG5 E2AH84 A0A1I8QAW2 A0A2A3E608 V9IC41 A0A0K8TNW3 A0A195DM10 A0A195BJA2 A0A158NA99 A0A195CBC0 A0A151X7K5 A0A026WK19 A0A087ZRF8 F4WN60 I5AMW9 A0A3B0K582 B4NGX2 A0A2M4CTV1 A0A0K8WG13 T1P9E3 A0A2M3ZJ06 B3MTA4 A0A0L0BNC7 A0A067RMC9 V5I6F0 A0A195FQZ9 A0A2M4ARC1 R4G8K7 A0A0R1EBL3 A0A146LTV8
Q9U6V6 A0A2H1WWQ8 S4PTE0 A0A2A4K1K7 A0A212EKW0 D6X3Z1 U5EUF7 A0A023EIU1 Q173S6 A0A1L8DUB4 A0A1Y1NH90 A0A1Q3G347 A0A1Q3G3A3 A0A1A9ZZK6 D3TRX1 W8BBJ0 A0A1B6MKP8 A0A034V3D6 A0A1I8QAY6 A0A182Q044 A0A0Q9X7A7 T1P818 A0A182W448 A0A1B6DPJ5 Q7QI07 A0A0K8UJK9 A0A0P8XUA6 A0A182RH62 A0A0A1X2A2 A0A154PFV2 A0A182JGC1 E2BFX9 B4HZS7 A0A084VN13 B4PMI8 Q9TVP3-2 H0RNN1 A0A182PPX2 B4R273 A0A182XNH9 A0A182V6C5 A0A182IDP0 A0A224XRB5 A0A069DPM1 B3P570 A0A0Q9WNV7 A0A023F8Q1 N6U8N6 A0A0Q9XAH2 A0A0N7Z9D6 T1HJG3 A0A182U6U9 A0A1W4WP47 A0A1A9VVH1 K7J003 A0A1W4W2P0 T1DS84 A0A182Y543 A0A1B0FAK0 A0A0J7NRY7 A0A2J7RPR6 A0A2M4C0W3 A0A336KZ16 A0A182FIG5 E2AH84 A0A1I8QAW2 A0A2A3E608 V9IC41 A0A0K8TNW3 A0A195DM10 A0A195BJA2 A0A158NA99 A0A195CBC0 A0A151X7K5 A0A026WK19 A0A087ZRF8 F4WN60 I5AMW9 A0A3B0K582 B4NGX2 A0A2M4CTV1 A0A0K8WG13 T1P9E3 A0A2M3ZJ06 B3MTA4 A0A0L0BNC7 A0A067RMC9 V5I6F0 A0A195FQZ9 A0A2M4ARC1 R4G8K7 A0A0R1EBL3 A0A146LTV8
Pubmed
26434795
10760603
19121390
26354079
23622113
22118469
+ More
18362917 19820115 24945155 17510324 28004739 20353571 24495485 25348373 17994087 25315136 12364791 14747013 17210077 25830018 20798317 24438588 17550304 10542335 10731132 12537572 12537569 26334808 25474469 23537049 27129103 20075255 25244985 26369729 21347285 24508170 30249741 21719571 15632085 26108605 24845553 26823975
18362917 19820115 24945155 17510324 28004739 20353571 24495485 25348373 17994087 25315136 12364791 14747013 17210077 25830018 20798317 24438588 17550304 10542335 10731132 12537572 12537569 26334808 25474469 23537049 27129103 20075255 25244985 26369729 21347285 24508170 30249741 21719571 15632085 26108605 24845553 26823975
EMBL
FJ592085
ACT34046.1
AF176014
JN872887
AFC01226.1
BABH01005689
+ More
KQ460883 KPJ11195.1 KQ459603 KPI92763.1 AF176015 ODYU01011634 SOQ57505.1 GAIX01012203 JAA80357.1 NWSH01000298 PCG77643.1 AGBW02014207 OWR42111.1 KQ971379 EEZ97348.1 GANO01001493 JAB58378.1 GAPW01005029 JAC08569.1 CH477417 EAT41309.1 GFDF01004061 JAV10023.1 GEZM01003098 JAV96918.1 GFDL01000821 JAV34224.1 GFDL01000750 JAV34295.1 EZ424173 ADD20449.1 GAMC01019331 GAMC01019328 JAB87224.1 GEBQ01023996 GEBQ01017808 GEBQ01017087 GEBQ01008773 GEBQ01004947 GEBQ01003493 JAT15981.1 JAT22169.1 JAT22890.1 JAT31204.1 JAT35030.1 JAT36484.1 GAKP01022305 JAC36647.1 AXCN02002023 CH964272 KRG00040.1 KA644781 AFP59410.1 GEDC01009693 JAS27605.1 AAAB01008811 EAA04936.1 GDHF01030423 GDHF01025774 GDHF01016135 JAI21891.1 JAI26540.1 JAI36179.1 CH902623 KPU72896.1 GBXI01008823 JAD05469.1 KQ434896 KZC10756.1 GL448096 EFN85381.1 CH480819 EDW53534.1 ATLV01014671 KE524978 KFB39357.1 CM000160 EDW99122.1 AF192462 AF192463 AF132905 AF132902 AF132903 AF132904 AE014297 AY060331 BT011341 BT132968 AEW48259.1 CM000364 EDX15029.1 APCN01004753 GFTR01002782 JAW13644.1 GBGD01003253 JAC85636.1 CH954182 EDV53053.1 CH940650 KRF83623.1 GBBI01000952 JAC17760.1 APGK01035409 APGK01035410 APGK01035411 KB740925 KB631924 ENN78030.1 ERL87275.1 CH933806 KRG01582.1 GDKW01000843 JAI55752.1 ACPB03007771 GAMD01001614 JAA99976.1 CCAG010006883 LBMM01002158 KMQ95200.1 NEVH01001355 PNF42824.1 GGFJ01009825 MBW58966.1 UFQS01001304 UFQT01001304 SSX10210.1 SSX29913.1 GL439483 EFN67201.1 KZ288375 PBC26676.1 JR037934 JR037935 AEY58041.1 AEY58042.1 GDAI01001765 JAI15838.1 KQ980734 KYN13871.1 KQ976455 KYM85189.1 ADTU01010084 KQ978009 KYM98159.1 KQ982446 KYQ56356.1 KK107167 QOIP01000010 EZA56397.1 RLU18037.1 GL888235 EGI64329.1 CM000070 EIM52304.2 OUUW01000005 SPP80101.1 EDW84469.1 GGFL01004080 MBW68258.1 GDHF01002206 JAI50108.1 KA644780 AFP59409.1 GGFM01007738 MBW28489.1 EDV30494.1 JRES01001702 KNC20774.1 KK852425 KDR24153.1 GALX01008300 JAB60166.1 KQ981305 KYN43010.1 GGFK01009998 MBW43319.1 GAHY01000802 JAA76708.1 KRK04666.1 GDHC01008074 JAQ10555.1
KQ460883 KPJ11195.1 KQ459603 KPI92763.1 AF176015 ODYU01011634 SOQ57505.1 GAIX01012203 JAA80357.1 NWSH01000298 PCG77643.1 AGBW02014207 OWR42111.1 KQ971379 EEZ97348.1 GANO01001493 JAB58378.1 GAPW01005029 JAC08569.1 CH477417 EAT41309.1 GFDF01004061 JAV10023.1 GEZM01003098 JAV96918.1 GFDL01000821 JAV34224.1 GFDL01000750 JAV34295.1 EZ424173 ADD20449.1 GAMC01019331 GAMC01019328 JAB87224.1 GEBQ01023996 GEBQ01017808 GEBQ01017087 GEBQ01008773 GEBQ01004947 GEBQ01003493 JAT15981.1 JAT22169.1 JAT22890.1 JAT31204.1 JAT35030.1 JAT36484.1 GAKP01022305 JAC36647.1 AXCN02002023 CH964272 KRG00040.1 KA644781 AFP59410.1 GEDC01009693 JAS27605.1 AAAB01008811 EAA04936.1 GDHF01030423 GDHF01025774 GDHF01016135 JAI21891.1 JAI26540.1 JAI36179.1 CH902623 KPU72896.1 GBXI01008823 JAD05469.1 KQ434896 KZC10756.1 GL448096 EFN85381.1 CH480819 EDW53534.1 ATLV01014671 KE524978 KFB39357.1 CM000160 EDW99122.1 AF192462 AF192463 AF132905 AF132902 AF132903 AF132904 AE014297 AY060331 BT011341 BT132968 AEW48259.1 CM000364 EDX15029.1 APCN01004753 GFTR01002782 JAW13644.1 GBGD01003253 JAC85636.1 CH954182 EDV53053.1 CH940650 KRF83623.1 GBBI01000952 JAC17760.1 APGK01035409 APGK01035410 APGK01035411 KB740925 KB631924 ENN78030.1 ERL87275.1 CH933806 KRG01582.1 GDKW01000843 JAI55752.1 ACPB03007771 GAMD01001614 JAA99976.1 CCAG010006883 LBMM01002158 KMQ95200.1 NEVH01001355 PNF42824.1 GGFJ01009825 MBW58966.1 UFQS01001304 UFQT01001304 SSX10210.1 SSX29913.1 GL439483 EFN67201.1 KZ288375 PBC26676.1 JR037934 JR037935 AEY58041.1 AEY58042.1 GDAI01001765 JAI15838.1 KQ980734 KYN13871.1 KQ976455 KYM85189.1 ADTU01010084 KQ978009 KYM98159.1 KQ982446 KYQ56356.1 KK107167 QOIP01000010 EZA56397.1 RLU18037.1 GL888235 EGI64329.1 CM000070 EIM52304.2 OUUW01000005 SPP80101.1 EDW84469.1 GGFL01004080 MBW68258.1 GDHF01002206 JAI50108.1 KA644780 AFP59409.1 GGFM01007738 MBW28489.1 EDV30494.1 JRES01001702 KNC20774.1 KK852425 KDR24153.1 GALX01008300 JAB60166.1 KQ981305 KYN43010.1 GGFK01009998 MBW43319.1 GAHY01000802 JAA76708.1 KRK04666.1 GDHC01008074 JAQ10555.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000007266
+ More
UP000008820 UP000092445 UP000095300 UP000075886 UP000007798 UP000095301 UP000075920 UP000007062 UP000007801 UP000075900 UP000076502 UP000075880 UP000008237 UP000001292 UP000030765 UP000002282 UP000000803 UP000075885 UP000000304 UP000076407 UP000075903 UP000075840 UP000008711 UP000008792 UP000019118 UP000030742 UP000009192 UP000015103 UP000075902 UP000192223 UP000078200 UP000002358 UP000192221 UP000076408 UP000092444 UP000036403 UP000235965 UP000069272 UP000000311 UP000242457 UP000078492 UP000078540 UP000005205 UP000078542 UP000075809 UP000053097 UP000279307 UP000005203 UP000007755 UP000001819 UP000268350 UP000037069 UP000027135 UP000078541
UP000008820 UP000092445 UP000095300 UP000075886 UP000007798 UP000095301 UP000075920 UP000007062 UP000007801 UP000075900 UP000076502 UP000075880 UP000008237 UP000001292 UP000030765 UP000002282 UP000000803 UP000075885 UP000000304 UP000076407 UP000075903 UP000075840 UP000008711 UP000008792 UP000019118 UP000030742 UP000009192 UP000015103 UP000075902 UP000192223 UP000078200 UP000002358 UP000192221 UP000076408 UP000092444 UP000036403 UP000235965 UP000069272 UP000000311 UP000242457 UP000078492 UP000078540 UP000005205 UP000078542 UP000075809 UP000053097 UP000279307 UP000005203 UP000007755 UP000001819 UP000268350 UP000037069 UP000027135 UP000078541
Pfam
PF00226 DnaJ
SUPFAM
SSF46565
SSF46565
Gene 3D
CDD
ProteinModelPortal
C7AR00
Q9U6V7
H6VTQ2
H9JLI6
A0A194R1S0
A0A194PI92
+ More
Q9U6V6 A0A2H1WWQ8 S4PTE0 A0A2A4K1K7 A0A212EKW0 D6X3Z1 U5EUF7 A0A023EIU1 Q173S6 A0A1L8DUB4 A0A1Y1NH90 A0A1Q3G347 A0A1Q3G3A3 A0A1A9ZZK6 D3TRX1 W8BBJ0 A0A1B6MKP8 A0A034V3D6 A0A1I8QAY6 A0A182Q044 A0A0Q9X7A7 T1P818 A0A182W448 A0A1B6DPJ5 Q7QI07 A0A0K8UJK9 A0A0P8XUA6 A0A182RH62 A0A0A1X2A2 A0A154PFV2 A0A182JGC1 E2BFX9 B4HZS7 A0A084VN13 B4PMI8 Q9TVP3-2 H0RNN1 A0A182PPX2 B4R273 A0A182XNH9 A0A182V6C5 A0A182IDP0 A0A224XRB5 A0A069DPM1 B3P570 A0A0Q9WNV7 A0A023F8Q1 N6U8N6 A0A0Q9XAH2 A0A0N7Z9D6 T1HJG3 A0A182U6U9 A0A1W4WP47 A0A1A9VVH1 K7J003 A0A1W4W2P0 T1DS84 A0A182Y543 A0A1B0FAK0 A0A0J7NRY7 A0A2J7RPR6 A0A2M4C0W3 A0A336KZ16 A0A182FIG5 E2AH84 A0A1I8QAW2 A0A2A3E608 V9IC41 A0A0K8TNW3 A0A195DM10 A0A195BJA2 A0A158NA99 A0A195CBC0 A0A151X7K5 A0A026WK19 A0A087ZRF8 F4WN60 I5AMW9 A0A3B0K582 B4NGX2 A0A2M4CTV1 A0A0K8WG13 T1P9E3 A0A2M3ZJ06 B3MTA4 A0A0L0BNC7 A0A067RMC9 V5I6F0 A0A195FQZ9 A0A2M4ARC1 R4G8K7 A0A0R1EBL3 A0A146LTV8
Q9U6V6 A0A2H1WWQ8 S4PTE0 A0A2A4K1K7 A0A212EKW0 D6X3Z1 U5EUF7 A0A023EIU1 Q173S6 A0A1L8DUB4 A0A1Y1NH90 A0A1Q3G347 A0A1Q3G3A3 A0A1A9ZZK6 D3TRX1 W8BBJ0 A0A1B6MKP8 A0A034V3D6 A0A1I8QAY6 A0A182Q044 A0A0Q9X7A7 T1P818 A0A182W448 A0A1B6DPJ5 Q7QI07 A0A0K8UJK9 A0A0P8XUA6 A0A182RH62 A0A0A1X2A2 A0A154PFV2 A0A182JGC1 E2BFX9 B4HZS7 A0A084VN13 B4PMI8 Q9TVP3-2 H0RNN1 A0A182PPX2 B4R273 A0A182XNH9 A0A182V6C5 A0A182IDP0 A0A224XRB5 A0A069DPM1 B3P570 A0A0Q9WNV7 A0A023F8Q1 N6U8N6 A0A0Q9XAH2 A0A0N7Z9D6 T1HJG3 A0A182U6U9 A0A1W4WP47 A0A1A9VVH1 K7J003 A0A1W4W2P0 T1DS84 A0A182Y543 A0A1B0FAK0 A0A0J7NRY7 A0A2J7RPR6 A0A2M4C0W3 A0A336KZ16 A0A182FIG5 E2AH84 A0A1I8QAW2 A0A2A3E608 V9IC41 A0A0K8TNW3 A0A195DM10 A0A195BJA2 A0A158NA99 A0A195CBC0 A0A151X7K5 A0A026WK19 A0A087ZRF8 F4WN60 I5AMW9 A0A3B0K582 B4NGX2 A0A2M4CTV1 A0A0K8WG13 T1P9E3 A0A2M3ZJ06 B3MTA4 A0A0L0BNC7 A0A067RMC9 V5I6F0 A0A195FQZ9 A0A2M4ARC1 R4G8K7 A0A0R1EBL3 A0A146LTV8
PDB
2CTQ
E-value=3.43297e-23,
Score=263
Ontologies
KEGG
PANTHER
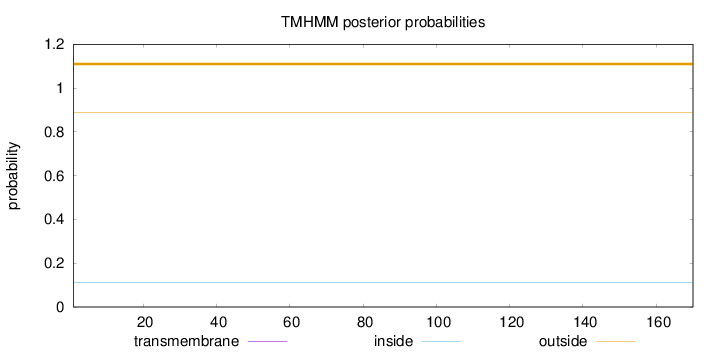
Topology
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11270
outside
1 - 170
Population Genetic Test Statistics
Pi
10.930328
Theta
12.19447
Tajima's D
-0.633752
CLR
23.443406
CSRT
0.215039248037598
Interpretation
Uncertain
