Pre Gene Modal
BGIBMGA010566
Annotation
prolyl_4-hydroxylase_alpha_subunit_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.183 Nuclear Reliability : 1.609
Sequence
CDS
ATGGATGACATCCCTAACTACCTCGGCAACCCGATCAATGCTTTCACGTTGATAAAAAGATTAACGGCCGACCTTGATTTTATAGAGGATAGCATTAAAATTGGAACAGAATACATAAAGAACGTCACAATGAACCACGTGGACGTGAAATATCCGTCATTGGAAGATCTGACGGGAGCAGCTCAGGCGCTGACTCGGTTGCAGGAAACCTACTATTTAAATGTACACGATCTAGCCGAGGGTATATTGAATGGAGTTTCATACAGTACTCCTATGACGGCGAGCGATTGTTACGAACTCGGCCGGACTCTATACAACGATAAAGATTACACAAACGCCTTGGCCTGGATGAAAGAAGCGCTAAGGAAATATAAAGATGAAAACGTCATGTACCCGTTCACCGAAGTCGATATCTTGGAATATATAGGATTCGCATATTATTTAAACGGAGACGTCAAAACCGCTCTGGAATGGACTCAGAGACTTCTGTCCGTCGATCCGAAGCACGTACGAGCGCGGGGCAACATACCGCACTATCAGAAGACGATAGCCGAACAAGAGGCCGAATTAAAGAAGCAACAACGTGGGGAAACCTCGGATGAGCCTGAAGAAGAAGATGGTCAAGATTACGAATTATCAGAGTACGCAAAGGAACGCAAAGTTTACGAATCGCTGTGTCGGGGAGAAATGGAAATACCCCATGAGATTACTAAGAGGTTGAAATGTTGGTACGTCACCGACACGCATCCGTTTTTAAAGTTGGCCCCAATCAAAGTGGAGCAGATGTACGTGAAGCCCGACATATTTATGTTCCACGAAGTGATGACCGACGACGAGATTGAGTTCATCAAAAAACGAGCAAAGCCGAGGTTCAAACGGGCTGTCGTTCACGACCCTAAAACTGGTGAGCTGACACCGGCCCATTACCGCATCAGCAAGTCGTCGTGGCTCCGCGACGAGGAGTCTCCGGTCATAGCGCGCATCACGCAGCGCGTCACCGACATGACCGGGCTCAGCATGCTGCACGCCGAGGAGCTTCAGGTCGTCAACTACGGCATAGGGGGACACTACGAACCGCACTTCGACTTCGCTAGGAAACGTGAGAATCCATTCACGAAATTCGGCGGCAACAGAATAGCCACCGTCCTCTTCTACATGTCTGACGTGGCGCAGGGCGGCGCTACAGTGTTCACCGAACTTGGACTCAGTTTGTTTCCAATAAAACGAGCTGCGGCGTTCTGGTTGAACCTGCACGCGTCGGGCGAAGGAGACCTCGCCACCAGGCATGCCGCCTGCCCCGTGCTCAGGGGATCCAAGTGGGTGTCAAATAAATGGATACATCAAGGCGGGCAAGAGCTGTTGAGGCCCTGCGACCTTGAGTACCAGGAGGAGGGCATCATCCGCAAGATTCCTCGTCCAGTGCCGAAGACATCCAGTACTTTAAAATTATCAAATATAAGTTTAAAAACTGTTCCTATAACTAGAAAATGCATGAAGAAATATAGCAAAGAAAACCCTCAAAAGACCTCTAAAGACCAGTGA
Protein
MDDIPNYLGNPINAFTLIKRLTADLDFIEDSIKIGTEYIKNVTMNHVDVKYPSLEDLTGAAQALTRLQETYYLNVHDLAEGILNGVSYSTPMTASDCYELGRTLYNDKDYTNALAWMKEALRKYKDENVMYPFTEVDILEYIGFAYYLNGDVKTALEWTQRLLSVDPKHVRARGNIPHYQKTIAEQEAELKKQQRGETSDEPEEEDGQDYELSEYAKERKVYESLCRGEMEIPHEITKRLKCWYVTDTHPFLKLAPIKVEQMYVKPDIFMFHEVMTDDEIEFIKKRAKPRFKRAVVHDPKTGELTPAHYRISKSSWLRDEESPVIARITQRVTDMTGLSMLHAEELQVVNYGIGGHYEPHFDFARKRENPFTKFGGNRIATVLFYMSDVAQGGATVFTELGLSLFPIKRAAAFWLNLHASGEGDLATRHAACPVLRGSKWVSNKWIHQGGQELLRPCDLEYQEEGIIRKIPRPVPKTSSTLKLSNISLKTVPITRKCMKKYSKENPQKTSKDQ
Summary
Uniprot
Q53EK0
A0A2A4K174
A0A2H1V446
A0A194PJ61
A0A3S2P863
A0A194R0G9
+ More
A0A194PH78 A0A182Y542 Q7PSX4 A0A2A3E5A6 B0XDL9 W5JLH9 A0A087ZR73 E1ZVX0 A0A1Q3FPJ6 A0A1Q3FP39 A0A1Q3FP59 A0A1Q3FP30 A0A182IDP1 A0A195AYN4 A0A195DYY7 A0A195F882 A0A023EWE6 A0A182XNH8 A0A182U7C8 A0A151WXN8 Q173S4 A0A1S4FFJ7 A0A158NP69 A0A067RK41 A0A084VN14 A0A026W8C1 A0A195CGD7 A0A182H720 A0A3L8DIF5 A0A0N0BE74 A0A0J7L862 A0A182MTX3 A0A154PH06 D6X3Y9 A0A182W447 A0A0A1X3X4 A0A034VA48 A0A182RL43 W8B8C2 A0A0K8VXA3 W8AYQ0 A0A1I8MY79 A0A1W4VQI1 T1PD32 A0A182FIG4 A0A1B6MIZ2 A0A182QSK5 B3MTB8 A0A182JID1 A0A3Q0IMV6 A0A1W4XI42 Q9VA69 B4K8R9 A0A0M4EPC7 B4PMH5 B4R1M7 A0A336MJ28 B4HZR4 A0A1J1I4Q8 B4LYM5 B3P583 U5EZI6 A0A336L025 A0A1I8Q4K5 A0A182MZX0 B4GNL7 Q29C45 A0A2J7RPT1 K7J002 A0A194R5Q5 A0A3B0JX82 A0A0C9PRJ6 B4JV99 A0A1B0CQR2 A0A1B6CXX7 E0VU49 A0A0P4Z620 B4NGW2 A0A0T6AZY8 E9G3W4 A0A0P5JGF9 A0A2K8JSJ0 A0A0P4YLU8 A0A0P5IUW0 A0A161N0P5 A0A0P4YFH3 A0A0N8AC92 A0A0P5KMC9 A0A0K8TML1 A0A0P5L1V0 A0A0P5ELI2 A0A0N8B8C6 A0A0P5JG06
A0A194PH78 A0A182Y542 Q7PSX4 A0A2A3E5A6 B0XDL9 W5JLH9 A0A087ZR73 E1ZVX0 A0A1Q3FPJ6 A0A1Q3FP39 A0A1Q3FP59 A0A1Q3FP30 A0A182IDP1 A0A195AYN4 A0A195DYY7 A0A195F882 A0A023EWE6 A0A182XNH8 A0A182U7C8 A0A151WXN8 Q173S4 A0A1S4FFJ7 A0A158NP69 A0A067RK41 A0A084VN14 A0A026W8C1 A0A195CGD7 A0A182H720 A0A3L8DIF5 A0A0N0BE74 A0A0J7L862 A0A182MTX3 A0A154PH06 D6X3Y9 A0A182W447 A0A0A1X3X4 A0A034VA48 A0A182RL43 W8B8C2 A0A0K8VXA3 W8AYQ0 A0A1I8MY79 A0A1W4VQI1 T1PD32 A0A182FIG4 A0A1B6MIZ2 A0A182QSK5 B3MTB8 A0A182JID1 A0A3Q0IMV6 A0A1W4XI42 Q9VA69 B4K8R9 A0A0M4EPC7 B4PMH5 B4R1M7 A0A336MJ28 B4HZR4 A0A1J1I4Q8 B4LYM5 B3P583 U5EZI6 A0A336L025 A0A1I8Q4K5 A0A182MZX0 B4GNL7 Q29C45 A0A2J7RPT1 K7J002 A0A194R5Q5 A0A3B0JX82 A0A0C9PRJ6 B4JV99 A0A1B0CQR2 A0A1B6CXX7 E0VU49 A0A0P4Z620 B4NGW2 A0A0T6AZY8 E9G3W4 A0A0P5JGF9 A0A2K8JSJ0 A0A0P4YLU8 A0A0P5IUW0 A0A161N0P5 A0A0P4YFH3 A0A0N8AC92 A0A0P5KMC9 A0A0K8TML1 A0A0P5L1V0 A0A0P5ELI2 A0A0N8B8C6 A0A0P5JG06
Pubmed
15890264
19121390
26354079
25244985
12364791
14747013
+ More
17210077 20920257 23761445 20798317 24945155 17510324 21347285 24845553 24438588 24508170 26483478 30249741 18362917 19820115 25830018 25348373 24495485 25315136 17994087 18057021 10731132 12537568 12537572 12537573 12537574 11850189 16110336 17569856 17569867 26109357 26109356 17550304 15632085 20075255 20566863 21292972 26369729
17210077 20920257 23761445 20798317 24945155 17510324 21347285 24845553 24438588 24508170 26483478 30249741 18362917 19820115 25830018 25348373 24495485 25315136 17994087 18057021 10731132 12537568 12537572 12537573 12537574 11850189 16110336 17569856 17569867 26109357 26109356 17550304 15632085 20075255 20566863 21292972 26369729
EMBL
BABH01005687
BABH01005688
BABH01005689
AY099068
AAM21932.1
NWSH01000298
+ More
PCG77644.1 ODYU01000375 SOQ35054.1 KQ459603 KPI92759.1 RSAL01000003 RVE54746.1 KQ460883 KPJ11197.1 KPI92761.1 AAAB01008811 EAA04901.5 KZ288375 PBC26674.1 DS232767 EDS45529.1 ADMH02001195 ETN63765.1 GL434663 EFN74662.1 GFDL01005637 JAV29408.1 GFDL01005763 JAV29282.1 GFDL01005670 JAV29375.1 GFDL01005694 JAV29351.1 APCN01004753 KQ976703 KYM77150.1 KQ980066 KYN17937.1 KQ981727 KYN36825.1 GAPW01000839 JAC12759.1 KQ982668 KYQ52537.1 CH477417 EAT41311.1 ADTU01022253 ADTU01022254 ADTU01022255 ADTU01022256 ADTU01022257 KK852425 KDR24152.1 ATLV01014671 KE524978 KFB39358.1 KK107347 EZA52305.1 KQ977791 KYM99809.1 JXUM01115432 JXUM01115433 KQ565879 KXJ70574.1 QOIP01000007 RLU20131.1 KQ435836 KOX71492.1 LBMM01000330 KMR00777.1 AXCM01009514 KQ434896 KZC10754.1 KQ971379 EEZ97484.1 GBXI01008944 JAD05348.1 GAKP01019972 GAKP01019971 GAKP01019970 JAC38982.1 GAMC01020541 JAB86014.1 GDHF01030537 GDHF01012446 GDHF01008817 GDHF01008803 GDHF01001872 JAI21777.1 JAI39868.1 JAI43497.1 JAI43511.1 JAI50442.1 GAMC01020539 JAB86016.1 KA645858 AFP60487.1 GEBQ01004104 JAT35873.1 AXCN02002023 CH902623 EDV30508.1 KPU72900.1 AE014297 AY052109 AF495536 AAF57053.2 AAK93533.1 AAM18058.1 CH933806 EDW15488.2 CP012526 ALC47627.1 CM000160 EDW99109.2 CM000364 EDX15015.1 UFQT01001304 SSX29915.1 CH480819 EDW53521.1 CVRI01000040 CRK94746.1 CH940650 EDW68045.2 CH954182 EDV53066.1 GANO01000729 JAB59142.1 UFQS01001467 UFQT01001467 SSX11130.1 SSX30701.1 CH479186 EDW39343.1 CM000070 EAL26796.1 NEVH01001355 PNF42840.1 KPJ11196.1 OUUW01000005 SPP80110.1 GBYB01003947 GBYB01003948 JAG73714.1 JAG73715.1 CH916374 EDV91419.1 AJWK01023887 GEDC01019014 JAS18284.1 AAZO01005441 DS235780 EEB16905.1 GDIP01229335 JAI94066.1 CH964272 EDW84459.1 LJIG01022453 KRT80516.1 GL732531 EFX85841.1 GDIQ01200303 JAK51422.1 KY031259 ATU83010.1 GDIP01229334 JAI94067.1 GDIQ01208806 GDIQ01207552 JAK44173.1 GEMB01002682 JAS00512.1 GDIP01229336 JAI94065.1 GDIP01161601 JAJ61801.1 GDIQ01208805 GDIQ01207550 GDIQ01202853 GDIP01089557 JAK44175.1 JAM14158.1 GDAI01002458 JAI15145.1 GDIQ01207551 GDIQ01183655 JAK68070.1 GDIP01144744 JAJ78658.1 GDIQ01202851 JAK48874.1 GDIQ01202852 JAK48873.1
PCG77644.1 ODYU01000375 SOQ35054.1 KQ459603 KPI92759.1 RSAL01000003 RVE54746.1 KQ460883 KPJ11197.1 KPI92761.1 AAAB01008811 EAA04901.5 KZ288375 PBC26674.1 DS232767 EDS45529.1 ADMH02001195 ETN63765.1 GL434663 EFN74662.1 GFDL01005637 JAV29408.1 GFDL01005763 JAV29282.1 GFDL01005670 JAV29375.1 GFDL01005694 JAV29351.1 APCN01004753 KQ976703 KYM77150.1 KQ980066 KYN17937.1 KQ981727 KYN36825.1 GAPW01000839 JAC12759.1 KQ982668 KYQ52537.1 CH477417 EAT41311.1 ADTU01022253 ADTU01022254 ADTU01022255 ADTU01022256 ADTU01022257 KK852425 KDR24152.1 ATLV01014671 KE524978 KFB39358.1 KK107347 EZA52305.1 KQ977791 KYM99809.1 JXUM01115432 JXUM01115433 KQ565879 KXJ70574.1 QOIP01000007 RLU20131.1 KQ435836 KOX71492.1 LBMM01000330 KMR00777.1 AXCM01009514 KQ434896 KZC10754.1 KQ971379 EEZ97484.1 GBXI01008944 JAD05348.1 GAKP01019972 GAKP01019971 GAKP01019970 JAC38982.1 GAMC01020541 JAB86014.1 GDHF01030537 GDHF01012446 GDHF01008817 GDHF01008803 GDHF01001872 JAI21777.1 JAI39868.1 JAI43497.1 JAI43511.1 JAI50442.1 GAMC01020539 JAB86016.1 KA645858 AFP60487.1 GEBQ01004104 JAT35873.1 AXCN02002023 CH902623 EDV30508.1 KPU72900.1 AE014297 AY052109 AF495536 AAF57053.2 AAK93533.1 AAM18058.1 CH933806 EDW15488.2 CP012526 ALC47627.1 CM000160 EDW99109.2 CM000364 EDX15015.1 UFQT01001304 SSX29915.1 CH480819 EDW53521.1 CVRI01000040 CRK94746.1 CH940650 EDW68045.2 CH954182 EDV53066.1 GANO01000729 JAB59142.1 UFQS01001467 UFQT01001467 SSX11130.1 SSX30701.1 CH479186 EDW39343.1 CM000070 EAL26796.1 NEVH01001355 PNF42840.1 KPJ11196.1 OUUW01000005 SPP80110.1 GBYB01003947 GBYB01003948 JAG73714.1 JAG73715.1 CH916374 EDV91419.1 AJWK01023887 GEDC01019014 JAS18284.1 AAZO01005441 DS235780 EEB16905.1 GDIP01229335 JAI94066.1 CH964272 EDW84459.1 LJIG01022453 KRT80516.1 GL732531 EFX85841.1 GDIQ01200303 JAK51422.1 KY031259 ATU83010.1 GDIP01229334 JAI94067.1 GDIQ01208806 GDIQ01207552 JAK44173.1 GEMB01002682 JAS00512.1 GDIP01229336 JAI94065.1 GDIP01161601 JAJ61801.1 GDIQ01208805 GDIQ01207550 GDIQ01202853 GDIP01089557 JAK44175.1 JAM14158.1 GDAI01002458 JAI15145.1 GDIQ01207551 GDIQ01183655 JAK68070.1 GDIP01144744 JAJ78658.1 GDIQ01202851 JAK48874.1 GDIQ01202852 JAK48873.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000076408
+ More
UP000007062 UP000242457 UP000002320 UP000000673 UP000005203 UP000000311 UP000075840 UP000078540 UP000078492 UP000078541 UP000076407 UP000075902 UP000075809 UP000008820 UP000005205 UP000027135 UP000030765 UP000053097 UP000078542 UP000069940 UP000249989 UP000279307 UP000053105 UP000036403 UP000075883 UP000076502 UP000007266 UP000075920 UP000075900 UP000095301 UP000192221 UP000069272 UP000075886 UP000007801 UP000075880 UP000079169 UP000192223 UP000000803 UP000009192 UP000092553 UP000002282 UP000000304 UP000001292 UP000183832 UP000008792 UP000008711 UP000095300 UP000075884 UP000008744 UP000001819 UP000235965 UP000002358 UP000268350 UP000001070 UP000092461 UP000009046 UP000007798 UP000000305
UP000007062 UP000242457 UP000002320 UP000000673 UP000005203 UP000000311 UP000075840 UP000078540 UP000078492 UP000078541 UP000076407 UP000075902 UP000075809 UP000008820 UP000005205 UP000027135 UP000030765 UP000053097 UP000078542 UP000069940 UP000249989 UP000279307 UP000053105 UP000036403 UP000075883 UP000076502 UP000007266 UP000075920 UP000075900 UP000095301 UP000192221 UP000069272 UP000075886 UP000007801 UP000075880 UP000079169 UP000192223 UP000000803 UP000009192 UP000092553 UP000002282 UP000000304 UP000001292 UP000183832 UP000008792 UP000008711 UP000095300 UP000075884 UP000008744 UP000001819 UP000235965 UP000002358 UP000268350 UP000001070 UP000092461 UP000009046 UP000007798 UP000000305
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
Q53EK0
A0A2A4K174
A0A2H1V446
A0A194PJ61
A0A3S2P863
A0A194R0G9
+ More
A0A194PH78 A0A182Y542 Q7PSX4 A0A2A3E5A6 B0XDL9 W5JLH9 A0A087ZR73 E1ZVX0 A0A1Q3FPJ6 A0A1Q3FP39 A0A1Q3FP59 A0A1Q3FP30 A0A182IDP1 A0A195AYN4 A0A195DYY7 A0A195F882 A0A023EWE6 A0A182XNH8 A0A182U7C8 A0A151WXN8 Q173S4 A0A1S4FFJ7 A0A158NP69 A0A067RK41 A0A084VN14 A0A026W8C1 A0A195CGD7 A0A182H720 A0A3L8DIF5 A0A0N0BE74 A0A0J7L862 A0A182MTX3 A0A154PH06 D6X3Y9 A0A182W447 A0A0A1X3X4 A0A034VA48 A0A182RL43 W8B8C2 A0A0K8VXA3 W8AYQ0 A0A1I8MY79 A0A1W4VQI1 T1PD32 A0A182FIG4 A0A1B6MIZ2 A0A182QSK5 B3MTB8 A0A182JID1 A0A3Q0IMV6 A0A1W4XI42 Q9VA69 B4K8R9 A0A0M4EPC7 B4PMH5 B4R1M7 A0A336MJ28 B4HZR4 A0A1J1I4Q8 B4LYM5 B3P583 U5EZI6 A0A336L025 A0A1I8Q4K5 A0A182MZX0 B4GNL7 Q29C45 A0A2J7RPT1 K7J002 A0A194R5Q5 A0A3B0JX82 A0A0C9PRJ6 B4JV99 A0A1B0CQR2 A0A1B6CXX7 E0VU49 A0A0P4Z620 B4NGW2 A0A0T6AZY8 E9G3W4 A0A0P5JGF9 A0A2K8JSJ0 A0A0P4YLU8 A0A0P5IUW0 A0A161N0P5 A0A0P4YFH3 A0A0N8AC92 A0A0P5KMC9 A0A0K8TML1 A0A0P5L1V0 A0A0P5ELI2 A0A0N8B8C6 A0A0P5JG06
A0A194PH78 A0A182Y542 Q7PSX4 A0A2A3E5A6 B0XDL9 W5JLH9 A0A087ZR73 E1ZVX0 A0A1Q3FPJ6 A0A1Q3FP39 A0A1Q3FP59 A0A1Q3FP30 A0A182IDP1 A0A195AYN4 A0A195DYY7 A0A195F882 A0A023EWE6 A0A182XNH8 A0A182U7C8 A0A151WXN8 Q173S4 A0A1S4FFJ7 A0A158NP69 A0A067RK41 A0A084VN14 A0A026W8C1 A0A195CGD7 A0A182H720 A0A3L8DIF5 A0A0N0BE74 A0A0J7L862 A0A182MTX3 A0A154PH06 D6X3Y9 A0A182W447 A0A0A1X3X4 A0A034VA48 A0A182RL43 W8B8C2 A0A0K8VXA3 W8AYQ0 A0A1I8MY79 A0A1W4VQI1 T1PD32 A0A182FIG4 A0A1B6MIZ2 A0A182QSK5 B3MTB8 A0A182JID1 A0A3Q0IMV6 A0A1W4XI42 Q9VA69 B4K8R9 A0A0M4EPC7 B4PMH5 B4R1M7 A0A336MJ28 B4HZR4 A0A1J1I4Q8 B4LYM5 B3P583 U5EZI6 A0A336L025 A0A1I8Q4K5 A0A182MZX0 B4GNL7 Q29C45 A0A2J7RPT1 K7J002 A0A194R5Q5 A0A3B0JX82 A0A0C9PRJ6 B4JV99 A0A1B0CQR2 A0A1B6CXX7 E0VU49 A0A0P4Z620 B4NGW2 A0A0T6AZY8 E9G3W4 A0A0P5JGF9 A0A2K8JSJ0 A0A0P4YLU8 A0A0P5IUW0 A0A161N0P5 A0A0P4YFH3 A0A0N8AC92 A0A0P5KMC9 A0A0K8TML1 A0A0P5L1V0 A0A0P5ELI2 A0A0N8B8C6 A0A0P5JG06
PDB
2V4A
E-value=5.97581e-29,
Score=319
Ontologies
KEGG
PATHWAY
GO
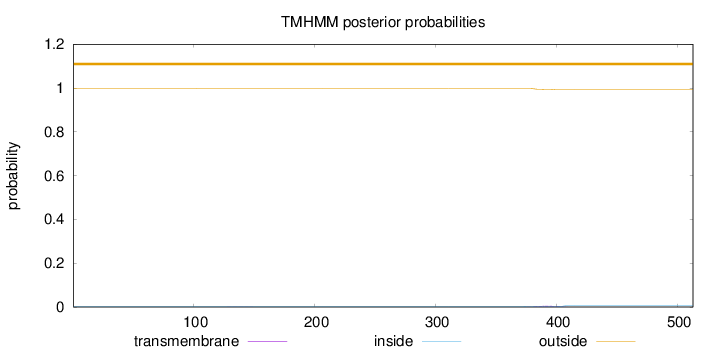
Topology
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11667
Exp number, first 60 AAs:
0.00095
Total prob of N-in:
0.00232
outside
1 - 513
Population Genetic Test Statistics
Pi
178.608739
Theta
140.598039
Tajima's D
-0.774375
CLR
1.065705
CSRT
0.180740962951852
Interpretation
Uncertain
