Pre Gene Modal
BGIBMGA010388
Annotation
PREDICTED:_DDB1-_and_CUL4-associated_factor_13_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.208
Sequence
CDS
ATGTCTAACGTAAAAATTAAAGTTATAAGTCGTAACCCAGAAGATTATTTGCGCGCTACGAAACGAGATATTCAGAAAATACCTCGCAACTATGACCCGAGCTTGCATCCACTAGAAGCACCCCGTGAATATGTGCGTGCTTTGAATGCTGTAAAATTAGAGAGGGTATTTGCCAAGCCCTTTATTGGTTGTTTAGATGGTCACAGAGATGGGGTTTCTACTATGGCCAAGCATCCTAGCCAACTAGCAGTTCTAGCTAGTGGAGCATTTGATGGTGAAGTACGTATTTGGGACTTAACTGTACGGAAGTGTACAAGAAATTTTGTTGCTCATGAGGGATGGGTGAGAGCTTTATGTTACAATCCAGATGGAAAGCATTTTATGAGTGTTGGTGATGATAAAACTATTAAAACGTGGAAAGCTGAAGCTGTCACAGAGGAAGATGAGGAACCCACTAATACGTTGCTAAGCATGTCAGTGGTGTCTGGTATAACACATCATAGAAACAAACCAATCTTTGCCACCTGTGGTGAACAATGTCAACTGTGGGAGAACACAAGAAATGAACCGATTAAAGTTTTTAAATGGGGCGTAGATAGCTTACATCATGTTTCATTTAACCAGGTTGAATACAATTTGTTGGCTTCTTGTGCTAGTGACAGAAGTATTATTTTATATGACTGCCGTGAATCTGGTCCTCTGAGGAAAGTTGTATTGGAGTTACGATCCAATGCACTCTCCTGGAACCCTATGGAAGCATTCATCTTTACAGTAGCTAATGAAGACTATAATTTGTACACATTTGATGTAAGGAGACTAAAAGAACCAGTGGCCATTCACATGGATCATACGTCTGCCGTGATTGATGTTGACTATGCGCCAACTGGCCGTGAATTTGTCGCGGGTAGCTATGACAAAACTATCAGAATATTCCAGAATGACAAAGGACACTCCCGAGATGTTTACCATACAAAAAGAATGCAGCGATTAACCTGCGTGAAATGGTCTCTAGATAATAAATATGTAATGAGTGGATCTGACGAAATGAATATAAGAATGTGGAAGGCCAGGGCATCTGAAAAACTTGGTGTGCTTAAGCCTCGTGAAAGACAAGCTCTTAATTATGCGGAAGCCCTAAAGGAGAAGTTTGCGACGCATCCACAAGTGAAGCGTATCGCTCGTCACAGACACGTCCCTAAACACATACTGAATGCGCAGAAAGAACTACGAACTATCCGAGAGAAAACGAAGAGAAAAGAAGCAAATAGACGCGCCCACAGTAAAAAGGGGACAATACCGGTTGTTTCTGAGAAAAAGAGACATGTCGTTAAAGAAGACGAATGA
Protein
MSNVKIKVISRNPEDYLRATKRDIQKIPRNYDPSLHPLEAPREYVRALNAVKLERVFAKPFIGCLDGHRDGVSTMAKHPSQLAVLASGAFDGEVRIWDLTVRKCTRNFVAHEGWVRALCYNPDGKHFMSVGDDKTIKTWKAEAVTEEDEEPTNTLLSMSVVSGITHHRNKPIFATCGEQCQLWENTRNEPIKVFKWGVDSLHHVSFNQVEYNLLASCASDRSIILYDCRESGPLRKVVLELRSNALSWNPMEAFIFTVANEDYNLYTFDVRRLKEPVAIHMDHTSAVIDVDYAPTGREFVAGSYDKTIRIFQNDKGHSRDVYHTKRMQRLTCVKWSLDNKYVMSGSDEMNIRMWKARASEKLGVLKPRERQALNYAEALKEKFATHPQVKRIARHRHVPKHILNAQKELRTIREKTKRKEANRRAHSKKGTIPVVSEKKRHVVKEDE
Summary
Similarity
Belongs to the peptidase C19 family.
Uniprot
H9JLI7
A0A194R5Q9
A0A194PHG8
A0A1E1WUS4
A0A2A4K0U9
A0A2H1V2I6
+ More
S4NIF0 A0A212EWA7 A0A3S2NRT3 D6W8S8 A0A087ZVV4 A0A2A3EFY4 A0A1Y1MYC5 A0A154PNE6 A0A0M9A2B5 A0A067QK63 A0A232ETH7 A0A2J7RPA9 A0A0L7RKT4 A0A158NYH8 A0A0C9PZ86 A0A195F641 A0A151ITA1 A0A195AYM4 A0A0J7KX70 A0A026WMM6 F4WT05 U4UL95 E2BL45 A0A151WHN7 A0A195CXI0 A0A310SJU2 A0A1B6E184 N6U300 A0A1W4W599 E2A657 A0A1I8NZH6 E9JD02 T1PAL9 A0A0A1XIB0 A0A1I8MNH7 A0A0L0C3P9 A0A034WMV6 A0A1L8DH58 W8BYE1 B7P621 A0A0K8UQD1 B4N502 A0A1W4VAK8 A0A1S3K776 A0A1S3K6T0 B3M714 B4L147 A0A1B0D8J4 A0A1B0G441 B4PCF6 Q9VUN7 B4ITZ7 A0A1A9XDT6 A0A1B0B8F7 I3IU87 A0A0J9RVH1 B4HI79 B3NI21 A0A3B4F4W9 A0A3Q4GYU1 B4LH67 D3TRG9 A0A1A9WGK1 A0A3P8R7Z3 A0A3P9C237 A0A3Q4BQZ0 A0A3Q2VR09 A0A0M5IYR0 L7M7G5 A0A1J1IUH4 A0A087U3V3 A0A224Z3N5 A0A3Q1FFH1 A0A3Q0R1L5 A0A131Z4K8 H3AZV2 B4IZ12 T1H724 A0A3B5BHV8 W5MTA1 A0A3Q1CB57 A0A3Q3IXZ5 A0A3S2PVM0 A0A131XLL4 A0A023FXF9 J9JM86 A0A023GK62 B4GUC4 A0A3B3D4F2 A0A3P9JWE8 A0A0K8TR14 A0A1W5ARW2 A0A3Q2ZHD4 E9H698
S4NIF0 A0A212EWA7 A0A3S2NRT3 D6W8S8 A0A087ZVV4 A0A2A3EFY4 A0A1Y1MYC5 A0A154PNE6 A0A0M9A2B5 A0A067QK63 A0A232ETH7 A0A2J7RPA9 A0A0L7RKT4 A0A158NYH8 A0A0C9PZ86 A0A195F641 A0A151ITA1 A0A195AYM4 A0A0J7KX70 A0A026WMM6 F4WT05 U4UL95 E2BL45 A0A151WHN7 A0A195CXI0 A0A310SJU2 A0A1B6E184 N6U300 A0A1W4W599 E2A657 A0A1I8NZH6 E9JD02 T1PAL9 A0A0A1XIB0 A0A1I8MNH7 A0A0L0C3P9 A0A034WMV6 A0A1L8DH58 W8BYE1 B7P621 A0A0K8UQD1 B4N502 A0A1W4VAK8 A0A1S3K776 A0A1S3K6T0 B3M714 B4L147 A0A1B0D8J4 A0A1B0G441 B4PCF6 Q9VUN7 B4ITZ7 A0A1A9XDT6 A0A1B0B8F7 I3IU87 A0A0J9RVH1 B4HI79 B3NI21 A0A3B4F4W9 A0A3Q4GYU1 B4LH67 D3TRG9 A0A1A9WGK1 A0A3P8R7Z3 A0A3P9C237 A0A3Q4BQZ0 A0A3Q2VR09 A0A0M5IYR0 L7M7G5 A0A1J1IUH4 A0A087U3V3 A0A224Z3N5 A0A3Q1FFH1 A0A3Q0R1L5 A0A131Z4K8 H3AZV2 B4IZ12 T1H724 A0A3B5BHV8 W5MTA1 A0A3Q1CB57 A0A3Q3IXZ5 A0A3S2PVM0 A0A131XLL4 A0A023FXF9 J9JM86 A0A023GK62 B4GUC4 A0A3B3D4F2 A0A3P9JWE8 A0A0K8TR14 A0A1W5ARW2 A0A3Q2ZHD4 E9H698
Pubmed
19121390
26354079
23622113
22118469
18362917
19820115
+ More
28004739 24845553 28648823 21347285 24508170 30249741 21719571 23537049 20798317 21282665 25830018 25315136 26108605 25348373 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25186727 22936249 20353571 25576852 28797301 26830274 9215903 28049606 29451363 17554307 26369729 21292972
28004739 24845553 28648823 21347285 24508170 30249741 21719571 23537049 20798317 21282665 25830018 25315136 26108605 25348373 24495485 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25186727 22936249 20353571 25576852 28797301 26830274 9215903 28049606 29451363 17554307 26369729 21292972
EMBL
BABH01005687
KQ460883
KPJ11201.1
KQ459603
KPI92757.1
GDQN01000284
+ More
JAT90770.1 NWSH01000298 PCG77646.1 ODYU01000375 SOQ35053.1 GAIX01014099 JAA78461.1 AGBW02012025 OWR45782.1 RSAL01000003 RVE54745.1 KQ971312 EEZ98265.1 KZ288262 PBC30394.1 GEZM01021529 JAV88956.1 KQ435004 KZC13401.1 KQ435775 KOX74996.1 KK853353 KDR08204.1 NNAY01002275 OXU21653.1 NEVH01001462 PNF42659.1 KQ414570 KOC71346.1 ADTU01003883 GBYB01006823 JAG76590.1 KQ981768 KYN35938.1 KQ981029 KYN10187.1 KQ976701 KYM77296.1 LBMM01002404 KMQ94869.1 KK107151 QOIP01000003 EZA57285.1 RLU25170.1 GL888331 EGI62618.1 KB632375 ERL93872.1 GL448921 EFN83595.1 KQ983114 KYQ47334.1 KQ977220 KYN04849.1 KQ760538 OAD59999.1 GEDC01005612 JAS31686.1 APGK01051684 KB741201 ENN72947.1 GL437108 EFN71116.1 GL771866 EFZ09142.1 KA644963 AFP59592.1 GBXI01003178 JAD11114.1 JRES01001015 KNC26114.1 GAKP01002061 JAC56891.1 GFDF01008400 JAV05684.1 GAMC01002193 JAC04363.1 ABJB010458834 DS643501 EEC02043.1 GDHF01023422 JAI28892.1 CH964101 EDW79441.1 CH902618 EDV40879.1 CH933809 EDW18204.1 AJVK01027480 CCAG010006632 CM000159 EDW94866.1 AE014296 AY058590 AAF49638.2 AAL13819.1 CH891752 EDW99860.1 JXJN01009933 AERX01031290 AERX01031291 AERX01031292 CM002912 KMY99741.1 CH480815 EDW41575.1 CH954178 EDV52107.1 CH940647 EDW70580.1 EZ424021 ADD20297.1 CP012525 ALC44048.1 GACK01004798 JAA60236.1 CVRI01000057 CRL02193.1 KK118034 KFM72042.1 GFPF01013342 MAA24488.1 GEDV01002825 JAP85732.1 AFYH01111405 AFYH01111406 AFYH01111407 AFYH01111408 CH916366 EDV97720.1 AHAT01004778 CM012452 RVE62242.1 GEFH01001553 JAP67028.1 GBBL01001151 JAC26169.1 ABLF02037926 GBBM01001104 JAC34314.1 CH479191 EDW26207.1 GDAI01000796 JAI16807.1 GL732597 EFX72618.1
JAT90770.1 NWSH01000298 PCG77646.1 ODYU01000375 SOQ35053.1 GAIX01014099 JAA78461.1 AGBW02012025 OWR45782.1 RSAL01000003 RVE54745.1 KQ971312 EEZ98265.1 KZ288262 PBC30394.1 GEZM01021529 JAV88956.1 KQ435004 KZC13401.1 KQ435775 KOX74996.1 KK853353 KDR08204.1 NNAY01002275 OXU21653.1 NEVH01001462 PNF42659.1 KQ414570 KOC71346.1 ADTU01003883 GBYB01006823 JAG76590.1 KQ981768 KYN35938.1 KQ981029 KYN10187.1 KQ976701 KYM77296.1 LBMM01002404 KMQ94869.1 KK107151 QOIP01000003 EZA57285.1 RLU25170.1 GL888331 EGI62618.1 KB632375 ERL93872.1 GL448921 EFN83595.1 KQ983114 KYQ47334.1 KQ977220 KYN04849.1 KQ760538 OAD59999.1 GEDC01005612 JAS31686.1 APGK01051684 KB741201 ENN72947.1 GL437108 EFN71116.1 GL771866 EFZ09142.1 KA644963 AFP59592.1 GBXI01003178 JAD11114.1 JRES01001015 KNC26114.1 GAKP01002061 JAC56891.1 GFDF01008400 JAV05684.1 GAMC01002193 JAC04363.1 ABJB010458834 DS643501 EEC02043.1 GDHF01023422 JAI28892.1 CH964101 EDW79441.1 CH902618 EDV40879.1 CH933809 EDW18204.1 AJVK01027480 CCAG010006632 CM000159 EDW94866.1 AE014296 AY058590 AAF49638.2 AAL13819.1 CH891752 EDW99860.1 JXJN01009933 AERX01031290 AERX01031291 AERX01031292 CM002912 KMY99741.1 CH480815 EDW41575.1 CH954178 EDV52107.1 CH940647 EDW70580.1 EZ424021 ADD20297.1 CP012525 ALC44048.1 GACK01004798 JAA60236.1 CVRI01000057 CRL02193.1 KK118034 KFM72042.1 GFPF01013342 MAA24488.1 GEDV01002825 JAP85732.1 AFYH01111405 AFYH01111406 AFYH01111407 AFYH01111408 CH916366 EDV97720.1 AHAT01004778 CM012452 RVE62242.1 GEFH01001553 JAP67028.1 GBBL01001151 JAC26169.1 ABLF02037926 GBBM01001104 JAC34314.1 CH479191 EDW26207.1 GDAI01000796 JAI16807.1 GL732597 EFX72618.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
+ More
UP000007266 UP000005203 UP000242457 UP000076502 UP000053105 UP000027135 UP000215335 UP000235965 UP000053825 UP000005205 UP000078541 UP000078492 UP000078540 UP000036403 UP000053097 UP000279307 UP000007755 UP000030742 UP000008237 UP000075809 UP000078542 UP000019118 UP000192223 UP000000311 UP000095300 UP000095301 UP000037069 UP000001555 UP000007798 UP000192221 UP000085678 UP000007801 UP000009192 UP000092462 UP000092444 UP000002282 UP000000803 UP000092443 UP000092460 UP000005207 UP000001292 UP000008711 UP000261460 UP000261580 UP000008792 UP000091820 UP000265100 UP000265160 UP000261620 UP000264840 UP000092553 UP000183832 UP000054359 UP000257200 UP000261340 UP000008672 UP000001070 UP000015102 UP000261400 UP000018468 UP000257160 UP000261600 UP000007819 UP000008744 UP000261560 UP000265180 UP000192224 UP000264800 UP000000305
UP000007266 UP000005203 UP000242457 UP000076502 UP000053105 UP000027135 UP000215335 UP000235965 UP000053825 UP000005205 UP000078541 UP000078492 UP000078540 UP000036403 UP000053097 UP000279307 UP000007755 UP000030742 UP000008237 UP000075809 UP000078542 UP000019118 UP000192223 UP000000311 UP000095300 UP000095301 UP000037069 UP000001555 UP000007798 UP000192221 UP000085678 UP000007801 UP000009192 UP000092462 UP000092444 UP000002282 UP000000803 UP000092443 UP000092460 UP000005207 UP000001292 UP000008711 UP000261460 UP000261580 UP000008792 UP000091820 UP000265100 UP000265160 UP000261620 UP000264840 UP000092553 UP000183832 UP000054359 UP000257200 UP000261340 UP000008672 UP000001070 UP000015102 UP000261400 UP000018468 UP000257160 UP000261600 UP000007819 UP000008744 UP000261560 UP000265180 UP000192224 UP000264800 UP000000305
Pfam
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR019775 WD40_repeat_CS
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR007287 Sof1
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR013979 TIF_beta_prop-like
IPR020472 G-protein_beta_WD-40_rep
IPR036623 Hemimethylated_DNA-bd_sf
IPR011722 Hemimethylated_DNA-bd_dom
IPR001394 Peptidase_C19_UCH
IPR036249 Thioredoxin-like_sf
IPR018200 USP_CS
IPR038765 Papain-like_cys_pep_sf
IPR013766 Thioredoxin_domain
IPR007474 ApaG_domain
IPR028889 USP_dom
IPR036767 ApaG_sf
IPR019775 WD40_repeat_CS
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR007287 Sof1
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR013979 TIF_beta_prop-like
IPR020472 G-protein_beta_WD-40_rep
IPR036623 Hemimethylated_DNA-bd_sf
IPR011722 Hemimethylated_DNA-bd_dom
IPR001394 Peptidase_C19_UCH
IPR036249 Thioredoxin-like_sf
IPR018200 USP_CS
IPR038765 Papain-like_cys_pep_sf
IPR013766 Thioredoxin_domain
IPR007474 ApaG_domain
IPR028889 USP_dom
IPR036767 ApaG_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JLI7
A0A194R5Q9
A0A194PHG8
A0A1E1WUS4
A0A2A4K0U9
A0A2H1V2I6
+ More
S4NIF0 A0A212EWA7 A0A3S2NRT3 D6W8S8 A0A087ZVV4 A0A2A3EFY4 A0A1Y1MYC5 A0A154PNE6 A0A0M9A2B5 A0A067QK63 A0A232ETH7 A0A2J7RPA9 A0A0L7RKT4 A0A158NYH8 A0A0C9PZ86 A0A195F641 A0A151ITA1 A0A195AYM4 A0A0J7KX70 A0A026WMM6 F4WT05 U4UL95 E2BL45 A0A151WHN7 A0A195CXI0 A0A310SJU2 A0A1B6E184 N6U300 A0A1W4W599 E2A657 A0A1I8NZH6 E9JD02 T1PAL9 A0A0A1XIB0 A0A1I8MNH7 A0A0L0C3P9 A0A034WMV6 A0A1L8DH58 W8BYE1 B7P621 A0A0K8UQD1 B4N502 A0A1W4VAK8 A0A1S3K776 A0A1S3K6T0 B3M714 B4L147 A0A1B0D8J4 A0A1B0G441 B4PCF6 Q9VUN7 B4ITZ7 A0A1A9XDT6 A0A1B0B8F7 I3IU87 A0A0J9RVH1 B4HI79 B3NI21 A0A3B4F4W9 A0A3Q4GYU1 B4LH67 D3TRG9 A0A1A9WGK1 A0A3P8R7Z3 A0A3P9C237 A0A3Q4BQZ0 A0A3Q2VR09 A0A0M5IYR0 L7M7G5 A0A1J1IUH4 A0A087U3V3 A0A224Z3N5 A0A3Q1FFH1 A0A3Q0R1L5 A0A131Z4K8 H3AZV2 B4IZ12 T1H724 A0A3B5BHV8 W5MTA1 A0A3Q1CB57 A0A3Q3IXZ5 A0A3S2PVM0 A0A131XLL4 A0A023FXF9 J9JM86 A0A023GK62 B4GUC4 A0A3B3D4F2 A0A3P9JWE8 A0A0K8TR14 A0A1W5ARW2 A0A3Q2ZHD4 E9H698
S4NIF0 A0A212EWA7 A0A3S2NRT3 D6W8S8 A0A087ZVV4 A0A2A3EFY4 A0A1Y1MYC5 A0A154PNE6 A0A0M9A2B5 A0A067QK63 A0A232ETH7 A0A2J7RPA9 A0A0L7RKT4 A0A158NYH8 A0A0C9PZ86 A0A195F641 A0A151ITA1 A0A195AYM4 A0A0J7KX70 A0A026WMM6 F4WT05 U4UL95 E2BL45 A0A151WHN7 A0A195CXI0 A0A310SJU2 A0A1B6E184 N6U300 A0A1W4W599 E2A657 A0A1I8NZH6 E9JD02 T1PAL9 A0A0A1XIB0 A0A1I8MNH7 A0A0L0C3P9 A0A034WMV6 A0A1L8DH58 W8BYE1 B7P621 A0A0K8UQD1 B4N502 A0A1W4VAK8 A0A1S3K776 A0A1S3K6T0 B3M714 B4L147 A0A1B0D8J4 A0A1B0G441 B4PCF6 Q9VUN7 B4ITZ7 A0A1A9XDT6 A0A1B0B8F7 I3IU87 A0A0J9RVH1 B4HI79 B3NI21 A0A3B4F4W9 A0A3Q4GYU1 B4LH67 D3TRG9 A0A1A9WGK1 A0A3P8R7Z3 A0A3P9C237 A0A3Q4BQZ0 A0A3Q2VR09 A0A0M5IYR0 L7M7G5 A0A1J1IUH4 A0A087U3V3 A0A224Z3N5 A0A3Q1FFH1 A0A3Q0R1L5 A0A131Z4K8 H3AZV2 B4IZ12 T1H724 A0A3B5BHV8 W5MTA1 A0A3Q1CB57 A0A3Q3IXZ5 A0A3S2PVM0 A0A131XLL4 A0A023FXF9 J9JM86 A0A023GK62 B4GUC4 A0A3B3D4F2 A0A3P9JWE8 A0A0K8TR14 A0A1W5ARW2 A0A3Q2ZHD4 E9H698
PDB
5WLC
E-value=2.51309e-105,
Score=977
Ontologies
GO
Topology
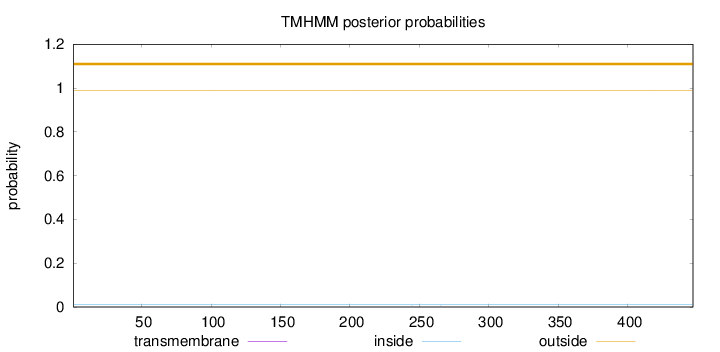
Length:
447
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000800000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00997
outside
1 - 447
Population Genetic Test Statistics
Pi
140.834486
Theta
120.250156
Tajima's D
0.701113
CLR
0.029311
CSRT
0.572171391430428
Interpretation
Uncertain
