Gene
KWMTBOMO07177 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010562
Annotation
PREDICTED:_striatin_isoform_X1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.367 Nuclear Reliability : 1.946
Sequence
CDS
ATGGATGATAACTCCGTATCTCATCACAACGGAGGGCAAGTGGGATCTCAAATTGGAGTCAGTGTAAATAATAAACAAAATGAAGAACCCAGTCAAAGTGTTCAGTATTCAATACCGGGCATTTTGCATTTTATCCAACACGAATGGGCGAGATTCGAACTGGAGAGATCACAATGGGAGGTGGACAAAGCAGAATTTGAGGCACGGATCGCCTTTCTCCAAGGTGAGAGGAAGGGTCAAGAAAATTTAAAAAATGATCTCGTGAGGCGGATTAAAATGCTTGAATATGCATTAAAGCAAGAAAGAGCAAAATTCCACAAGCTCAAGTATGGAGTGGAGTTGCAACAAGGAGACGTAAGGCCCCCGCCGGAAGAGTCAGCGACAGAGCCCGAACCAGTGGAACGCACGCAATGGAAGCAGGGAAGACAATTTATAAAACAGTACTTGCAGGAAATAGGCTACACGGATACAATTCTGGACGTGCGCTCTAATAGGGTACGCGCTCTCCTCGGTTTGAAGAACGAGGATTCAGATAACTCGAACTACAGAAACTATGAAAAACAACAGCACGACAGATGCGATTACTCTGCGAACCATATGGTTCGCAAATATGACTATGGTAAAGAGCCACGCAAAACGAGCGGTTCGTCTGGAGGCGGTTACAATGATGAAGGGATGTCCGTACAGGAGACAGCTGCAGTATTCGCTAACTTCGAGTTCTTGACCAATCAAGAAATGGATATGGACGAAATTGACGATCTCGATGCCAAACAGACTCTTCACGCTTCTGGTAAACAGGGTGAAGAGGTCGATCATGAAGCAGAAGAAGTTTTGAATGAGCTTAATATGCTTACTATAAATGAAGTCGTAAGTGCTGGCCAGGGGGATGAGTATCAAGGTGGAGTAACGAGCGGCGGCAGTGCGAGTGCGAGTGCGGCGTCGAGCAGCGTCGGCAGCGGGAGCGGCGCCGGGGAGGCTGACGAGGAGACGCTCGGACTCGGAGAGCTTGCCCAGCTCACCGTCAGCAACGAGCCCGAAGCTTATGATGTTGCCGGCGCTAATAAAGAATCTTTCCGCAAGACGTGGAACGCCAAATACACGCTCCGCTCGCACTTTGATGGGGTCCGCGCTATCGCATTCCATCCGACAGAAGCGGCGCTGGTGACAGCATCCGAGGATCACACATTGAAGTTATGGAACTTGCAACGTACCGTACCTGCGAAGAGATCGGCTGGTCTGGATGTGGAGCCACTGTACACGTTCCGCGCGCACTCAGCGCCCGTGCTCTGTCTCGCCATGGGGGCGCCGCGCTCTGAGGAATGCTTCTCCGGAGGGCTCGACGGCACCATTCGAGTCTGGAACCTACCTCCTCCCACCGCTGACCCGTACGATCCGTATGATCCCGCTGTCCAGGGTGCGGTTCTACGCGAACACACGGATGCGGTGTGGTCGCTGGCGAGTGCGTGCGGACGACTGGCGTCTGCATCGGCGGACGGCACGGTGCGGCTGTGGGCGCCGCGAGCCCCGCGCCCCCTGCTGGCGACGCTGCGTCCGGACGACGGCCCGCCCCCCGCCGCCGGGCCCGCCTTGGCGCCCTCGCCCGCGCCCGCTGCTCTCGACTTTGCCGACGCAGCCTCGCGAGCCGCAGTCGCTTACCGCGACGGATCTCTTCTGCTCTACGACCTTGAGACTGGCCAGGCCGTGTTGCGGTTGGGTTGCGACAGTCCCGCGAACCGCGTTCGTTCTCATCCGACGCTGCCGATGTTGGTGACCGCGCACGAGGACCGACACATTCGGTTCTGGGACGCGGTCTCGGGGCAGTGCTCGCACGCGATGGTGGCCCACTTGGACGCCGTCACGGGACTCGCTCTCGATCCCAACGGCCTGTTCCTGCTATCGGGCTCGCACGACTGCTCCGTGCGCCTCTGGAATCTCGACACGAAGACTTGCGTGCAGGAGATCACCGCGCACCGCAAGAAGTTCGACGAAAGCATCCTCGACGTCGCCTTCCACCCGTTGCGGCCGTACATCGCTAGTGCGGGCGCCGACGGTCTCGCCAAAGTATTCGTCTGA
Protein
MDDNSVSHHNGGQVGSQIGVSVNNKQNEEPSQSVQYSIPGILHFIQHEWARFELERSQWEVDKAEFEARIAFLQGERKGQENLKNDLVRRIKMLEYALKQERAKFHKLKYGVELQQGDVRPPPEESATEPEPVERTQWKQGRQFIKQYLQEIGYTDTILDVRSNRVRALLGLKNEDSDNSNYRNYEKQQHDRCDYSANHMVRKYDYGKEPRKTSGSSGGGYNDEGMSVQETAAVFANFEFLTNQEMDMDEIDDLDAKQTLHASGKQGEEVDHEAEEVLNELNMLTINEVVSAGQGDEYQGGVTSGGSASASAASSSVGSGSGAGEADEETLGLGELAQLTVSNEPEAYDVAGANKESFRKTWNAKYTLRSHFDGVRAIAFHPTEAALVTASEDHTLKLWNLQRTVPAKRSAGLDVEPLYTFRAHSAPVLCLAMGAPRSEECFSGGLDGTIRVWNLPPPTADPYDPYDPAVQGAVLREHTDAVWSLASACGRLASASADGTVRLWAPRAPRPLLATLRPDDGPPPAAGPALAPSPAPAALDFADAASRAAVAYRDGSLLLYDLETGQAVLRLGCDSPANRVRSHPTLPMLVTAHEDRHIRFWDAVSGQCSHAMVAHLDAVTGLALDPNGLFLLSGSHDCSVRLWNLDTKTCVQEITAHRKKFDESILDVAFHPLRPYIASAGADGLAKVFV
Summary
Uniprot
H9JM10
A0A2H1VCR1
A0A2A4JZJ0
A0A194PI77
A0A194R119
A0A212EPL0
+ More
A0A2J7PMR0 D2A379 A0A067RB71 A0A1B6GEK5 A0A1B6L3Z5 A0A1B6KUB9 A0A1Y1NC86 A0A146M3K7 A0A1B6DPZ4 A0A151JTH3 A0A0A9XRX4 A0A151IY21 F4W3Z0 A0A195CJX0 A0A232F9R1 E2APP1 A0A151X1D0 A0A158NTD9 A0A195B3U5 K7IPR7 V9IC70 A0A182YM17 A0A0K8TSG0 A0A224XKL5 A0A088AAE5 A0A0V0GA14 A0A2L2YC45 E2BKP6 A0A2A3E2F9 A0A1L8DXR1 U4UFJ6 A0A023F2U1 A0A2M3Z3A1 A0A3F2YQ63 A0A2M4CRA9 A0A1L8DXA7 A0A224YV07 A0A131YUX5 A0A2C9GQ85 A0A1S4GZ53 A0A087TF71 E9I9R1 A0A2M3ZGJ0 A0A2M4A2X9 A0A0C9R1K6 A0A2M4BF89 A0A2M3Z3E9 A0A2M4CSN6 A0A1E1XLB8 A0A1Q3F507 A0A293MZM5 A0A1Q3F5A9 Q5TS28 A0A084VMP2 A0A034WEC2 A0A1Q3F599 A0A0A1WW61 A0A336LKD5 A0A1I8P0K7 A0A0R3NT29 A0A1I8P0L3 A0A0P8XET0 A0A0R3NZ13 A0A0K8UID1 A0A0K8VRE8 A0A0P8XF23 A0A034WCT5 A0A1I8P0J8 A0A0A1XMI3 A0A0R1DRW7 A0A0Q5WKC0 A0A0J9TID4 B4NWC6 B3N6Z3 B4HYD6 A0A0J9QY72 D3DML3 A0A3B3SRG6 A0A3Q2I8K1 A0A146ZM85 A0A3B4ZYP9 A0A3Q0S0B1 A0A3Q1IK73 A0A3B4UCG7 A0A087YKL4 A0A3Q3G580 A0A3P8T3F5 A0A1A7XRM9 H2TV49 A0A3Q3K1W6 A0A3P9B7G1 A0A3Q1B1Y2 A0A3Q3FEF8
A0A2J7PMR0 D2A379 A0A067RB71 A0A1B6GEK5 A0A1B6L3Z5 A0A1B6KUB9 A0A1Y1NC86 A0A146M3K7 A0A1B6DPZ4 A0A151JTH3 A0A0A9XRX4 A0A151IY21 F4W3Z0 A0A195CJX0 A0A232F9R1 E2APP1 A0A151X1D0 A0A158NTD9 A0A195B3U5 K7IPR7 V9IC70 A0A182YM17 A0A0K8TSG0 A0A224XKL5 A0A088AAE5 A0A0V0GA14 A0A2L2YC45 E2BKP6 A0A2A3E2F9 A0A1L8DXR1 U4UFJ6 A0A023F2U1 A0A2M3Z3A1 A0A3F2YQ63 A0A2M4CRA9 A0A1L8DXA7 A0A224YV07 A0A131YUX5 A0A2C9GQ85 A0A1S4GZ53 A0A087TF71 E9I9R1 A0A2M3ZGJ0 A0A2M4A2X9 A0A0C9R1K6 A0A2M4BF89 A0A2M3Z3E9 A0A2M4CSN6 A0A1E1XLB8 A0A1Q3F507 A0A293MZM5 A0A1Q3F5A9 Q5TS28 A0A084VMP2 A0A034WEC2 A0A1Q3F599 A0A0A1WW61 A0A336LKD5 A0A1I8P0K7 A0A0R3NT29 A0A1I8P0L3 A0A0P8XET0 A0A0R3NZ13 A0A0K8UID1 A0A0K8VRE8 A0A0P8XF23 A0A034WCT5 A0A1I8P0J8 A0A0A1XMI3 A0A0R1DRW7 A0A0Q5WKC0 A0A0J9TID4 B4NWC6 B3N6Z3 B4HYD6 A0A0J9QY72 D3DML3 A0A3B3SRG6 A0A3Q2I8K1 A0A146ZM85 A0A3B4ZYP9 A0A3Q0S0B1 A0A3Q1IK73 A0A3B4UCG7 A0A087YKL4 A0A3Q3G580 A0A3P8T3F5 A0A1A7XRM9 H2TV49 A0A3Q3K1W6 A0A3P9B7G1 A0A3Q1B1Y2 A0A3Q3FEF8
Pubmed
19121390
26354079
22118469
18362917
19820115
24845553
+ More
28004739 26823975 25401762 21719571 28648823 20798317 21347285 20075255 25244985 26369729 26561354 23537049 25474469 28797301 26830274 21282665 29209593 12364791 24438588 25348373 25830018 15632085 17994087 23185243 18057021 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29240929 19892987 21551351 25186727
28004739 26823975 25401762 21719571 28648823 20798317 21347285 20075255 25244985 26369729 26561354 23537049 25474469 28797301 26830274 21282665 29209593 12364791 24438588 25348373 25830018 15632085 17994087 23185243 18057021 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29240929 19892987 21551351 25186727
EMBL
BABH01005659
ODYU01001839
SOQ38591.1
NWSH01000345
PCG77229.1
KQ459603
+ More
KPI92748.1 KQ460883 KPJ11209.1 AGBW02013442 OWR43432.1 NEVH01023960 PNF17621.1 KQ971338 EFA01950.2 KK852572 KDR21101.1 GECZ01008905 JAS60864.1 GEBQ01026434 GEBQ01021538 GEBQ01012024 JAT13543.1 JAT18439.1 JAT27953.1 GEBQ01024940 GEBQ01015106 GEBQ01013243 JAT15037.1 JAT24871.1 JAT26734.1 GEZM01006997 JAV95493.1 GDHC01004286 JAQ14343.1 GEDC01025028 GEDC01024780 GEDC01009567 JAS12270.1 JAS12518.1 JAS27731.1 KQ981885 KYN33866.1 GBHO01022018 GBHO01022016 GBHO01022015 JAG21586.1 JAG21588.1 JAG21589.1 KQ980778 KYN13213.1 GL887491 EGI70986.1 KQ977642 KYN01025.1 NNAY01000577 OXU27586.1 GL441572 EFN64592.1 KQ982585 KYQ54223.1 ADTU01025849 ADTU01025850 KQ976625 KYM78945.1 JR039466 AEY58718.1 GDAI01000306 JAI17297.1 GFTR01007867 JAW08559.1 GECL01001363 JAP04761.1 IAAA01013954 LAA04940.1 GL448819 EFN83744.1 KZ288444 PBC25664.1 GFDF01003039 JAV11045.1 KB632166 ERL89331.1 GBBI01003062 JAC15650.1 GGFM01002245 MBW22996.1 GGFL01003684 MBW67862.1 GFDF01003040 JAV11044.1 GFPF01006658 MAA17804.1 GEDV01005780 JAP82777.1 APCN01000644 KK114949 KFM63760.1 GL761846 EFZ22677.1 GGFM01006936 MBW27687.1 GGFK01001845 MBW35166.1 GBYB01006737 JAG76504.1 GGFJ01002561 MBW51702.1 GGFM01002298 MBW23049.1 GGFL01004097 MBW68275.1 GFAA01003406 JAU00029.1 GFDL01012398 JAV22647.1 GFWV01020660 MAA45388.1 GFDL01012387 JAV22658.1 AAAB01008944 EAL40293.3 ATLV01014616 KE524975 KFB39236.1 GAKP01006457 JAC52495.1 GFDL01012397 JAV22648.1 GBXI01011451 JAD02841.1 UFQT01000040 SSX18664.1 CH379060 KRT04092.1 CH902620 KPU73347.1 KRT04094.1 KRT04096.1 KRT04097.1 KRT04098.1 KRT04099.1 GDHF01031926 GDHF01027555 GDHF01026021 GDHF01022898 GDHF01021380 GDHF01012717 GDHF01009805 GDHF01006097 GDHF01002015 JAI20388.1 JAI24759.1 JAI26293.1 JAI29416.1 JAI30934.1 JAI39597.1 JAI42509.1 JAI46217.1 JAI50299.1 GDHF01011169 JAI41145.1 KPU73345.1 KPU73346.1 KPU73348.1 KPU73349.1 KPU73351.1 KPU73352.1 GAKP01006458 JAC52494.1 GBXI01015598 GBXI01006025 GBXI01002212 JAC98693.1 JAD08267.1 JAD12080.1 CM000157 KRJ97785.1 CH954177 KQS70731.1 CM002910 KMY88905.1 EDW88443.1 KRJ97781.1 KRJ97782.1 KRJ97783.1 KRJ97784.1 KRJ97786.1 KRJ97788.1 EDV59289.1 KQS70727.1 KQS70728.1 KQS70729.1 KQS70733.1 KQS70734.1 CH480818 EDW52066.1 KMY88902.1 KMY88907.1 KMY88908.1 BT120179 AE014134 ADB91427.1 AFH03610.1 GCES01018844 JAR67479.1 AYCK01018986 HADW01019045 HADX01003539 SBP20445.1
KPI92748.1 KQ460883 KPJ11209.1 AGBW02013442 OWR43432.1 NEVH01023960 PNF17621.1 KQ971338 EFA01950.2 KK852572 KDR21101.1 GECZ01008905 JAS60864.1 GEBQ01026434 GEBQ01021538 GEBQ01012024 JAT13543.1 JAT18439.1 JAT27953.1 GEBQ01024940 GEBQ01015106 GEBQ01013243 JAT15037.1 JAT24871.1 JAT26734.1 GEZM01006997 JAV95493.1 GDHC01004286 JAQ14343.1 GEDC01025028 GEDC01024780 GEDC01009567 JAS12270.1 JAS12518.1 JAS27731.1 KQ981885 KYN33866.1 GBHO01022018 GBHO01022016 GBHO01022015 JAG21586.1 JAG21588.1 JAG21589.1 KQ980778 KYN13213.1 GL887491 EGI70986.1 KQ977642 KYN01025.1 NNAY01000577 OXU27586.1 GL441572 EFN64592.1 KQ982585 KYQ54223.1 ADTU01025849 ADTU01025850 KQ976625 KYM78945.1 JR039466 AEY58718.1 GDAI01000306 JAI17297.1 GFTR01007867 JAW08559.1 GECL01001363 JAP04761.1 IAAA01013954 LAA04940.1 GL448819 EFN83744.1 KZ288444 PBC25664.1 GFDF01003039 JAV11045.1 KB632166 ERL89331.1 GBBI01003062 JAC15650.1 GGFM01002245 MBW22996.1 GGFL01003684 MBW67862.1 GFDF01003040 JAV11044.1 GFPF01006658 MAA17804.1 GEDV01005780 JAP82777.1 APCN01000644 KK114949 KFM63760.1 GL761846 EFZ22677.1 GGFM01006936 MBW27687.1 GGFK01001845 MBW35166.1 GBYB01006737 JAG76504.1 GGFJ01002561 MBW51702.1 GGFM01002298 MBW23049.1 GGFL01004097 MBW68275.1 GFAA01003406 JAU00029.1 GFDL01012398 JAV22647.1 GFWV01020660 MAA45388.1 GFDL01012387 JAV22658.1 AAAB01008944 EAL40293.3 ATLV01014616 KE524975 KFB39236.1 GAKP01006457 JAC52495.1 GFDL01012397 JAV22648.1 GBXI01011451 JAD02841.1 UFQT01000040 SSX18664.1 CH379060 KRT04092.1 CH902620 KPU73347.1 KRT04094.1 KRT04096.1 KRT04097.1 KRT04098.1 KRT04099.1 GDHF01031926 GDHF01027555 GDHF01026021 GDHF01022898 GDHF01021380 GDHF01012717 GDHF01009805 GDHF01006097 GDHF01002015 JAI20388.1 JAI24759.1 JAI26293.1 JAI29416.1 JAI30934.1 JAI39597.1 JAI42509.1 JAI46217.1 JAI50299.1 GDHF01011169 JAI41145.1 KPU73345.1 KPU73346.1 KPU73348.1 KPU73349.1 KPU73351.1 KPU73352.1 GAKP01006458 JAC52494.1 GBXI01015598 GBXI01006025 GBXI01002212 JAC98693.1 JAD08267.1 JAD12080.1 CM000157 KRJ97785.1 CH954177 KQS70731.1 CM002910 KMY88905.1 EDW88443.1 KRJ97781.1 KRJ97782.1 KRJ97783.1 KRJ97784.1 KRJ97786.1 KRJ97788.1 EDV59289.1 KQS70727.1 KQS70728.1 KQS70729.1 KQS70733.1 KQS70734.1 CH480818 EDW52066.1 KMY88902.1 KMY88907.1 KMY88908.1 BT120179 AE014134 ADB91427.1 AFH03610.1 GCES01018844 JAR67479.1 AYCK01018986 HADW01019045 HADX01003539 SBP20445.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000235965
+ More
UP000007266 UP000027135 UP000078541 UP000078492 UP000007755 UP000078542 UP000215335 UP000000311 UP000075809 UP000005205 UP000078540 UP000002358 UP000076408 UP000005203 UP000008237 UP000242457 UP000030742 UP000069272 UP000075840 UP000054359 UP000007062 UP000030765 UP000095300 UP000001819 UP000007801 UP000002282 UP000008711 UP000001292 UP000000803 UP000261540 UP000002281 UP000261400 UP000261340 UP000265040 UP000261420 UP000028760 UP000261660 UP000265080 UP000005226 UP000261600 UP000265160 UP000257160 UP000264800
UP000007266 UP000027135 UP000078541 UP000078492 UP000007755 UP000078542 UP000215335 UP000000311 UP000075809 UP000005205 UP000078540 UP000002358 UP000076408 UP000005203 UP000008237 UP000242457 UP000030742 UP000069272 UP000075840 UP000054359 UP000007062 UP000030765 UP000095300 UP000001819 UP000007801 UP000002282 UP000008711 UP000001292 UP000000803 UP000261540 UP000002281 UP000261400 UP000261340 UP000265040 UP000261420 UP000028760 UP000261660 UP000265080 UP000005226 UP000261600 UP000265160 UP000257160 UP000264800
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR001680 WD40_repeat
IPR013258 Striatin_N
IPR017986 WD40_repeat_dom
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR041489 PDZ_6
IPR001478 PDZ
IPR036034 PDZ_sf
IPR001680 WD40_repeat
IPR013258 Striatin_N
IPR017986 WD40_repeat_dom
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR041489 PDZ_6
IPR001478 PDZ
IPR036034 PDZ_sf
Gene 3D
ProteinModelPortal
H9JM10
A0A2H1VCR1
A0A2A4JZJ0
A0A194PI77
A0A194R119
A0A212EPL0
+ More
A0A2J7PMR0 D2A379 A0A067RB71 A0A1B6GEK5 A0A1B6L3Z5 A0A1B6KUB9 A0A1Y1NC86 A0A146M3K7 A0A1B6DPZ4 A0A151JTH3 A0A0A9XRX4 A0A151IY21 F4W3Z0 A0A195CJX0 A0A232F9R1 E2APP1 A0A151X1D0 A0A158NTD9 A0A195B3U5 K7IPR7 V9IC70 A0A182YM17 A0A0K8TSG0 A0A224XKL5 A0A088AAE5 A0A0V0GA14 A0A2L2YC45 E2BKP6 A0A2A3E2F9 A0A1L8DXR1 U4UFJ6 A0A023F2U1 A0A2M3Z3A1 A0A3F2YQ63 A0A2M4CRA9 A0A1L8DXA7 A0A224YV07 A0A131YUX5 A0A2C9GQ85 A0A1S4GZ53 A0A087TF71 E9I9R1 A0A2M3ZGJ0 A0A2M4A2X9 A0A0C9R1K6 A0A2M4BF89 A0A2M3Z3E9 A0A2M4CSN6 A0A1E1XLB8 A0A1Q3F507 A0A293MZM5 A0A1Q3F5A9 Q5TS28 A0A084VMP2 A0A034WEC2 A0A1Q3F599 A0A0A1WW61 A0A336LKD5 A0A1I8P0K7 A0A0R3NT29 A0A1I8P0L3 A0A0P8XET0 A0A0R3NZ13 A0A0K8UID1 A0A0K8VRE8 A0A0P8XF23 A0A034WCT5 A0A1I8P0J8 A0A0A1XMI3 A0A0R1DRW7 A0A0Q5WKC0 A0A0J9TID4 B4NWC6 B3N6Z3 B4HYD6 A0A0J9QY72 D3DML3 A0A3B3SRG6 A0A3Q2I8K1 A0A146ZM85 A0A3B4ZYP9 A0A3Q0S0B1 A0A3Q1IK73 A0A3B4UCG7 A0A087YKL4 A0A3Q3G580 A0A3P8T3F5 A0A1A7XRM9 H2TV49 A0A3Q3K1W6 A0A3P9B7G1 A0A3Q1B1Y2 A0A3Q3FEF8
A0A2J7PMR0 D2A379 A0A067RB71 A0A1B6GEK5 A0A1B6L3Z5 A0A1B6KUB9 A0A1Y1NC86 A0A146M3K7 A0A1B6DPZ4 A0A151JTH3 A0A0A9XRX4 A0A151IY21 F4W3Z0 A0A195CJX0 A0A232F9R1 E2APP1 A0A151X1D0 A0A158NTD9 A0A195B3U5 K7IPR7 V9IC70 A0A182YM17 A0A0K8TSG0 A0A224XKL5 A0A088AAE5 A0A0V0GA14 A0A2L2YC45 E2BKP6 A0A2A3E2F9 A0A1L8DXR1 U4UFJ6 A0A023F2U1 A0A2M3Z3A1 A0A3F2YQ63 A0A2M4CRA9 A0A1L8DXA7 A0A224YV07 A0A131YUX5 A0A2C9GQ85 A0A1S4GZ53 A0A087TF71 E9I9R1 A0A2M3ZGJ0 A0A2M4A2X9 A0A0C9R1K6 A0A2M4BF89 A0A2M3Z3E9 A0A2M4CSN6 A0A1E1XLB8 A0A1Q3F507 A0A293MZM5 A0A1Q3F5A9 Q5TS28 A0A084VMP2 A0A034WEC2 A0A1Q3F599 A0A0A1WW61 A0A336LKD5 A0A1I8P0K7 A0A0R3NT29 A0A1I8P0L3 A0A0P8XET0 A0A0R3NZ13 A0A0K8UID1 A0A0K8VRE8 A0A0P8XF23 A0A034WCT5 A0A1I8P0J8 A0A0A1XMI3 A0A0R1DRW7 A0A0Q5WKC0 A0A0J9TID4 B4NWC6 B3N6Z3 B4HYD6 A0A0J9QY72 D3DML3 A0A3B3SRG6 A0A3Q2I8K1 A0A146ZM85 A0A3B4ZYP9 A0A3Q0S0B1 A0A3Q1IK73 A0A3B4UCG7 A0A087YKL4 A0A3Q3G580 A0A3P8T3F5 A0A1A7XRM9 H2TV49 A0A3Q3K1W6 A0A3P9B7G1 A0A3Q1B1Y2 A0A3Q3FEF8
PDB
2YMU
E-value=3.22545e-19,
Score=236
Ontologies
GO
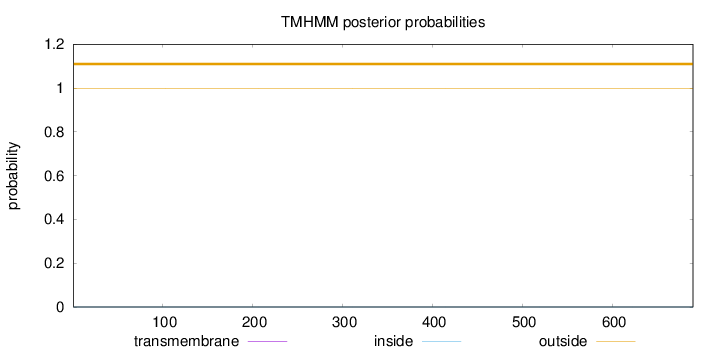
Topology
Length:
690
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0026
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00045
outside
1 - 690
Population Genetic Test Statistics
Pi
199.685052
Theta
153.962437
Tajima's D
-0.773561
CLR
100.244791
CSRT
0.178591070446478
Interpretation
Uncertain
