Gene
KWMTBOMO07176 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010396
Annotation
uncharacterized_protein_LOC106123913_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.927
Sequence
CDS
ATGTCACGCTATGGTGACTGCAAAGTTTATGTTGGAGATTTGGGGAATAATGCTAGTAAACCTGAACTTGAAGATGCCTTTTCATATTATGGTCCACTGAGGAATGTGTGGGTTGCAAGAAATCCTCCAGGCTTCGCATTTGTTGAATTCGAAGATCCACGGGATGCTGAAGACGCAGTACGTGGTTTGGATGGACGGACGATTTGCGGTCGTAGAGCCCGTGTAGAAATGTCTAATGGGGGTCGCGGTTATGGCAGCCGGGGTCCACCACCTCGTTCGCGTCTGCCTCCTCGGCCGTATGATGATCGTTGTTACGACTGCGGCGACAGAGGACACTACGCTCGTGACTGTTCACGTCGTCGGCGACGCAGGTTATCATTGGGTTGTGCGCCGAGAACGAACTCGTCAGTGTCTGCACTTCTTATTATCTGCGCTACGTAG
Protein
MSRYGDCKVYVGDLGNNASKPELEDAFSYYGPLRNVWVARNPPGFAFVEFEDPRDAEDAVRGLDGRTICGRRARVEMSNGGRGYGSRGPPPRSRLPPRPYDDRCYDCGDRGHYARDCSRRRRRRLSLGCAPRTNSSVSALLIICAT
Summary
Uniprot
A0A2A4K0E8
A0A1E1WME9
I4DJK9
A0A194R1T3
H9JLJ5
A0A212EPP6
+ More
A0A2A4JZ06 A0A2A4K018 A0A0L7KNE8 Q1HPK8 S4P5H1 A0A194PHF9 S4P8J1 A0A1W4XVV3 A0A1W4XWV9 A0A1L8DFV3 A0A1Y1K8Z4 A0A1Y1K8Y3 N6UEV3 A0A1B0DQU0 D6X2L1 A0A1B6FAP4 A0A1B6HX66 E2BJ42 J9JRS0 A0A1B6L7I2 E0VJB3 A0A2H8TE44 A0A2S2PR98 A0A232EXB5 A0A2A3EIN9 E2AE95 A0A336MA05 A0A0J7NZA5 A0A3L8E149 A0A310SQZ9 A0A0L7RJU4 D2DST7 A0A0C9QR32 A0A0A9WIZ2 A0A0A9W8N1 A0A0A9YSY6 A0A0A9WAK8 A0A0A9YXC2 A0A0C9PPJ3 A0A0A9WE70 A0A087ZPW7 R4V1F6 B1VK45 A0A161MAD8 A0A151WS07 A0A151HZY3 A0A158NE21 A0A151ISJ3 A0A151JX57 A0A224XSZ1 A0A0P4VHC3 A0A3R7PCB5 A0A182M133 E0VL84 E9J0U5 A0A0V0G8N3 A0A023F4S2 A0A182Q8Q1 A0A151III9 T1I507 A0A0P5Z661 A0A182NU70 A0A0P6EWQ7 A0A0P5ZAP8 A0A1J1I482 A0A3P8V759 A0A0P5ITJ7 A0A3Q2SZ23 A0A3P9Q9R4 A0A182W6N2 A0A3P9Q8Y8 A0A182Y3E6 A0A182RMD1 A0A182PAU8 A0A0P5AA72 A0A0P6DRG3 A0A3Q3VYD7 A0A0P5I8J1 A0A3B5LX61 A0A3B5B908 A0A3B5BG09 A0A023FXP6 A0A0P5QKT5 A0A2P6KR79 A0A3B5B902 A0A2M4CQZ8 A0A0P5VN20 A0A224YMV6 A0A1E1X1Q1 V5I0Y3 A0A0P5SS81 A0A224Z769 A0A131Z1Y1 A0A3Q0R2I9
A0A2A4JZ06 A0A2A4K018 A0A0L7KNE8 Q1HPK8 S4P5H1 A0A194PHF9 S4P8J1 A0A1W4XVV3 A0A1W4XWV9 A0A1L8DFV3 A0A1Y1K8Z4 A0A1Y1K8Y3 N6UEV3 A0A1B0DQU0 D6X2L1 A0A1B6FAP4 A0A1B6HX66 E2BJ42 J9JRS0 A0A1B6L7I2 E0VJB3 A0A2H8TE44 A0A2S2PR98 A0A232EXB5 A0A2A3EIN9 E2AE95 A0A336MA05 A0A0J7NZA5 A0A3L8E149 A0A310SQZ9 A0A0L7RJU4 D2DST7 A0A0C9QR32 A0A0A9WIZ2 A0A0A9W8N1 A0A0A9YSY6 A0A0A9WAK8 A0A0A9YXC2 A0A0C9PPJ3 A0A0A9WE70 A0A087ZPW7 R4V1F6 B1VK45 A0A161MAD8 A0A151WS07 A0A151HZY3 A0A158NE21 A0A151ISJ3 A0A151JX57 A0A224XSZ1 A0A0P4VHC3 A0A3R7PCB5 A0A182M133 E0VL84 E9J0U5 A0A0V0G8N3 A0A023F4S2 A0A182Q8Q1 A0A151III9 T1I507 A0A0P5Z661 A0A182NU70 A0A0P6EWQ7 A0A0P5ZAP8 A0A1J1I482 A0A3P8V759 A0A0P5ITJ7 A0A3Q2SZ23 A0A3P9Q9R4 A0A182W6N2 A0A3P9Q8Y8 A0A182Y3E6 A0A182RMD1 A0A182PAU8 A0A0P5AA72 A0A0P6DRG3 A0A3Q3VYD7 A0A0P5I8J1 A0A3B5LX61 A0A3B5B908 A0A3B5BG09 A0A023FXP6 A0A0P5QKT5 A0A2P6KR79 A0A3B5B902 A0A2M4CQZ8 A0A0P5VN20 A0A224YMV6 A0A1E1X1Q1 V5I0Y3 A0A0P5SS81 A0A224Z769 A0A131Z1Y1 A0A3Q0R2I9
Pubmed
EMBL
NWSH01000345
PCG77243.1
GDQN01002899
JAT88155.1
AK401477
BAM18099.1
+ More
KQ460883 KPJ11210.1 BABH01005659 AGBW02013442 OWR43431.1 PCG77245.1 PCG77244.1 JTDY01008585 KOB64499.1 DQ443394 DQ497207 ABF51483.1 ABF55972.2 GAIX01005549 JAA87011.1 KQ459603 KPI92747.1 GAIX01005906 JAA86654.1 GFDF01008870 JAV05214.1 GEZM01088780 GEZM01088779 GEZM01088770 GEZM01088769 GEZM01088764 GEZM01088762 JAV57932.1 GEZM01088783 GEZM01088781 GEZM01088773 GEZM01088765 JAV57922.1 APGK01029461 APGK01029462 APGK01029463 KB740735 ENN79141.1 AJVK01008748 KQ971372 EFA09429.1 GECZ01022531 JAS47238.1 GECU01028450 JAS79256.1 GL448558 EFN84192.1 ABLF02027459 GEBQ01020330 JAT19647.1 DS235221 EEB13469.1 GFXV01000187 MBW11992.1 GGMR01019370 MBY31989.1 NNAY01001755 OXU23021.1 KZ288238 PBC31334.1 GL438827 EFN68328.1 UFQT01000770 SSX27106.1 LBMM01000694 KMQ97725.1 QOIP01000001 RLU26434.1 KQ760832 OAD58886.1 KQ414579 KOC71109.1 FJ774776 ACY66497.1 GBYB01003122 JAG72889.1 GBHO01038779 GBRD01008394 JAG04825.1 JAG57427.1 GBHO01038782 JAG04822.1 GBHO01009401 GBRD01006452 JAG34203.1 JAG59369.1 GBHO01038780 JAG04824.1 GBHO01009399 GBRD01006451 JAG34205.1 JAG59370.1 GBYB01003123 JAG72890.1 GBHO01038781 JAG04823.1 KC571978 AGM32477.1 AM950284 CAQ34906.1 GEMB01007011 JAR96355.1 KQ982796 KYQ50593.1 KQ976687 KYM78145.1 ADTU01013114 ADTU01013115 KQ981064 KYN09842.1 KQ981607 KYN39417.1 GFTR01002212 JAW14214.1 GDKW01003586 JAI53009.1 QCYY01000931 ROT81713.1 AXCM01013990 DS235271 EEB14140.1 GL767538 EFZ13615.1 GECL01002381 JAP03743.1 GBBI01002295 JAC16417.1 AXCN02000773 KQ977477 KYN02486.1 ACPB03010532 GDIP01048425 JAM55290.1 GDIQ01057024 JAN37713.1 GDIP01046826 JAM56889.1 CVRI01000038 CRK94404.1 GDIQ01209919 JAK41806.1 GDIP01214350 JAJ09052.1 GDIQ01074665 JAN20072.1 GDIQ01223512 JAK28213.1 GBBL01000679 JAC26641.1 GDIQ01119052 GDIP01056573 JAL32674.1 JAM47142.1 MWRG01006646 PRD28816.1 GGFL01003423 MBW67601.1 GDIP01097817 JAM05898.1 GFPF01005933 MAA17079.1 GFAC01005988 JAT93200.1 GANP01001661 JAB82807.1 GDIP01136060 JAL67654.1 GFPF01012045 MAA23191.1 GEDV01003812 JAP84745.1
KQ460883 KPJ11210.1 BABH01005659 AGBW02013442 OWR43431.1 PCG77245.1 PCG77244.1 JTDY01008585 KOB64499.1 DQ443394 DQ497207 ABF51483.1 ABF55972.2 GAIX01005549 JAA87011.1 KQ459603 KPI92747.1 GAIX01005906 JAA86654.1 GFDF01008870 JAV05214.1 GEZM01088780 GEZM01088779 GEZM01088770 GEZM01088769 GEZM01088764 GEZM01088762 JAV57932.1 GEZM01088783 GEZM01088781 GEZM01088773 GEZM01088765 JAV57922.1 APGK01029461 APGK01029462 APGK01029463 KB740735 ENN79141.1 AJVK01008748 KQ971372 EFA09429.1 GECZ01022531 JAS47238.1 GECU01028450 JAS79256.1 GL448558 EFN84192.1 ABLF02027459 GEBQ01020330 JAT19647.1 DS235221 EEB13469.1 GFXV01000187 MBW11992.1 GGMR01019370 MBY31989.1 NNAY01001755 OXU23021.1 KZ288238 PBC31334.1 GL438827 EFN68328.1 UFQT01000770 SSX27106.1 LBMM01000694 KMQ97725.1 QOIP01000001 RLU26434.1 KQ760832 OAD58886.1 KQ414579 KOC71109.1 FJ774776 ACY66497.1 GBYB01003122 JAG72889.1 GBHO01038779 GBRD01008394 JAG04825.1 JAG57427.1 GBHO01038782 JAG04822.1 GBHO01009401 GBRD01006452 JAG34203.1 JAG59369.1 GBHO01038780 JAG04824.1 GBHO01009399 GBRD01006451 JAG34205.1 JAG59370.1 GBYB01003123 JAG72890.1 GBHO01038781 JAG04823.1 KC571978 AGM32477.1 AM950284 CAQ34906.1 GEMB01007011 JAR96355.1 KQ982796 KYQ50593.1 KQ976687 KYM78145.1 ADTU01013114 ADTU01013115 KQ981064 KYN09842.1 KQ981607 KYN39417.1 GFTR01002212 JAW14214.1 GDKW01003586 JAI53009.1 QCYY01000931 ROT81713.1 AXCM01013990 DS235271 EEB14140.1 GL767538 EFZ13615.1 GECL01002381 JAP03743.1 GBBI01002295 JAC16417.1 AXCN02000773 KQ977477 KYN02486.1 ACPB03010532 GDIP01048425 JAM55290.1 GDIQ01057024 JAN37713.1 GDIP01046826 JAM56889.1 CVRI01000038 CRK94404.1 GDIQ01209919 JAK41806.1 GDIP01214350 JAJ09052.1 GDIQ01074665 JAN20072.1 GDIQ01223512 JAK28213.1 GBBL01000679 JAC26641.1 GDIQ01119052 GDIP01056573 JAL32674.1 JAM47142.1 MWRG01006646 PRD28816.1 GGFL01003423 MBW67601.1 GDIP01097817 JAM05898.1 GFPF01005933 MAA17079.1 GFAC01005988 JAT93200.1 GANP01001661 JAB82807.1 GDIP01136060 JAL67654.1 GFPF01012045 MAA23191.1 GEDV01003812 JAP84745.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000007151
UP000037510
UP000053268
+ More
UP000192223 UP000019118 UP000092462 UP000007266 UP000008237 UP000007819 UP000009046 UP000215335 UP000242457 UP000000311 UP000036403 UP000279307 UP000053825 UP000005203 UP000075809 UP000078540 UP000005205 UP000078492 UP000078541 UP000283509 UP000075883 UP000075886 UP000078542 UP000015103 UP000075884 UP000183832 UP000265120 UP000265000 UP000242638 UP000075920 UP000076408 UP000075900 UP000075885 UP000261620 UP000261380 UP000261400 UP000261340
UP000192223 UP000019118 UP000092462 UP000007266 UP000008237 UP000007819 UP000009046 UP000215335 UP000242457 UP000000311 UP000036403 UP000279307 UP000053825 UP000005203 UP000075809 UP000078540 UP000005205 UP000078492 UP000078541 UP000283509 UP000075883 UP000075886 UP000078542 UP000015103 UP000075884 UP000183832 UP000265120 UP000265000 UP000242638 UP000075920 UP000076408 UP000075900 UP000075885 UP000261620 UP000261380 UP000261400 UP000261340
Interpro
IPR000504
RRM_dom
+ More
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001878 Znf_CCHC
IPR035979 RBD_domain_sf
IPR035022 PI3kinase_P85_nSH2
IPR000980 SH2
IPR032498 PI3K_P85_iSH2
IPR036860 SH2_dom_sf
IPR036875 Znf_CCHC_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001878 Znf_CCHC
IPR035979 RBD_domain_sf
IPR035022 PI3kinase_P85_nSH2
IPR000980 SH2
IPR032498 PI3K_P85_iSH2
IPR036860 SH2_dom_sf
IPR036875 Znf_CCHC_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
Gene 3D
ProteinModelPortal
A0A2A4K0E8
A0A1E1WME9
I4DJK9
A0A194R1T3
H9JLJ5
A0A212EPP6
+ More
A0A2A4JZ06 A0A2A4K018 A0A0L7KNE8 Q1HPK8 S4P5H1 A0A194PHF9 S4P8J1 A0A1W4XVV3 A0A1W4XWV9 A0A1L8DFV3 A0A1Y1K8Z4 A0A1Y1K8Y3 N6UEV3 A0A1B0DQU0 D6X2L1 A0A1B6FAP4 A0A1B6HX66 E2BJ42 J9JRS0 A0A1B6L7I2 E0VJB3 A0A2H8TE44 A0A2S2PR98 A0A232EXB5 A0A2A3EIN9 E2AE95 A0A336MA05 A0A0J7NZA5 A0A3L8E149 A0A310SQZ9 A0A0L7RJU4 D2DST7 A0A0C9QR32 A0A0A9WIZ2 A0A0A9W8N1 A0A0A9YSY6 A0A0A9WAK8 A0A0A9YXC2 A0A0C9PPJ3 A0A0A9WE70 A0A087ZPW7 R4V1F6 B1VK45 A0A161MAD8 A0A151WS07 A0A151HZY3 A0A158NE21 A0A151ISJ3 A0A151JX57 A0A224XSZ1 A0A0P4VHC3 A0A3R7PCB5 A0A182M133 E0VL84 E9J0U5 A0A0V0G8N3 A0A023F4S2 A0A182Q8Q1 A0A151III9 T1I507 A0A0P5Z661 A0A182NU70 A0A0P6EWQ7 A0A0P5ZAP8 A0A1J1I482 A0A3P8V759 A0A0P5ITJ7 A0A3Q2SZ23 A0A3P9Q9R4 A0A182W6N2 A0A3P9Q8Y8 A0A182Y3E6 A0A182RMD1 A0A182PAU8 A0A0P5AA72 A0A0P6DRG3 A0A3Q3VYD7 A0A0P5I8J1 A0A3B5LX61 A0A3B5B908 A0A3B5BG09 A0A023FXP6 A0A0P5QKT5 A0A2P6KR79 A0A3B5B902 A0A2M4CQZ8 A0A0P5VN20 A0A224YMV6 A0A1E1X1Q1 V5I0Y3 A0A0P5SS81 A0A224Z769 A0A131Z1Y1 A0A3Q0R2I9
A0A2A4JZ06 A0A2A4K018 A0A0L7KNE8 Q1HPK8 S4P5H1 A0A194PHF9 S4P8J1 A0A1W4XVV3 A0A1W4XWV9 A0A1L8DFV3 A0A1Y1K8Z4 A0A1Y1K8Y3 N6UEV3 A0A1B0DQU0 D6X2L1 A0A1B6FAP4 A0A1B6HX66 E2BJ42 J9JRS0 A0A1B6L7I2 E0VJB3 A0A2H8TE44 A0A2S2PR98 A0A232EXB5 A0A2A3EIN9 E2AE95 A0A336MA05 A0A0J7NZA5 A0A3L8E149 A0A310SQZ9 A0A0L7RJU4 D2DST7 A0A0C9QR32 A0A0A9WIZ2 A0A0A9W8N1 A0A0A9YSY6 A0A0A9WAK8 A0A0A9YXC2 A0A0C9PPJ3 A0A0A9WE70 A0A087ZPW7 R4V1F6 B1VK45 A0A161MAD8 A0A151WS07 A0A151HZY3 A0A158NE21 A0A151ISJ3 A0A151JX57 A0A224XSZ1 A0A0P4VHC3 A0A3R7PCB5 A0A182M133 E0VL84 E9J0U5 A0A0V0G8N3 A0A023F4S2 A0A182Q8Q1 A0A151III9 T1I507 A0A0P5Z661 A0A182NU70 A0A0P6EWQ7 A0A0P5ZAP8 A0A1J1I482 A0A3P8V759 A0A0P5ITJ7 A0A3Q2SZ23 A0A3P9Q9R4 A0A182W6N2 A0A3P9Q8Y8 A0A182Y3E6 A0A182RMD1 A0A182PAU8 A0A0P5AA72 A0A0P6DRG3 A0A3Q3VYD7 A0A0P5I8J1 A0A3B5LX61 A0A3B5B908 A0A3B5BG09 A0A023FXP6 A0A0P5QKT5 A0A2P6KR79 A0A3B5B902 A0A2M4CQZ8 A0A0P5VN20 A0A224YMV6 A0A1E1X1Q1 V5I0Y3 A0A0P5SS81 A0A224Z769 A0A131Z1Y1 A0A3Q0R2I9
PDB
2I38
E-value=6.239e-31,
Score=328
Ontologies
GO
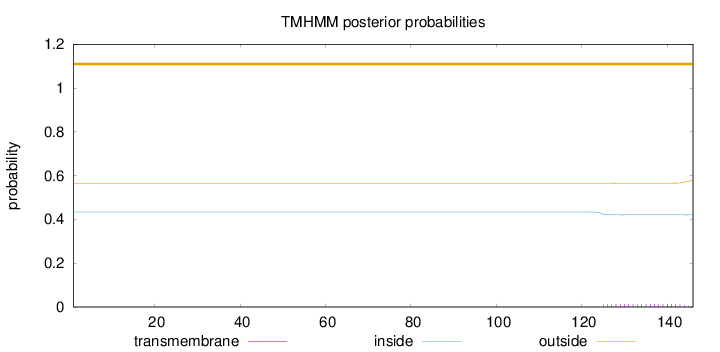
Topology
Length:
146
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26863
Exp number, first 60 AAs:
0
Total prob of N-in:
0.43444
outside
1 - 146
Population Genetic Test Statistics
Pi
120.336172
Theta
146.647073
Tajima's D
-0.72057
CLR
0.115381
CSRT
0.192340382980851
Interpretation
Uncertain
