Gene
KWMTBOMO07175
Pre Gene Modal
BGIBMGA010561
Annotation
phosphatidylinositol_3-kinase_60_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.288 Nuclear Reliability : 2.263
Sequence
CDS
ATGGTCGAAATAGGAGTTCCACAGCAGTCACCAGAACATAGCCTAGAAGCTGCGGAGTGGTACTGGGGCGATATAACAAGAGATGAAGCCAATGAAAAACTTCGAGACACTCGTGATGGAACTTTTCTTGTTCGTAATGCATCTAATAAGGATAGTGGATATACATTGACAGTCAGGAAAGGTGGTGCAAATAAGTTGATCAAAATATACCATCATGAAGGTCGATATGGCTTCCGTGAACCTTTTGAATTTAAGTCAGTAGTTGATCTTGTATGCTATTACAGTGAATACTCCCTGGCAACATGTAACAGTAGTCTAGACATCAAACTCTTGTATCCATTGTCTAGACATCAAGAAAATGAAGGAGGTGAAAACATAAGTGATGAAACTTTAGCAACTAGGTATAAAGAGATACATAAGAAGTATATATTAAAAACTAGGGATTACAATATAAGATTTTCTAAATATACAAGATTAAGGGAAGAAGTTACTTTAAAACGACAAGCTTTAGAAGCGTTTAGAGAAGCTGTAAAAATGTGTGAAGACCACATTAAACTACAAGAAAAAATACAAGCAGAAGCACAGCCTCATGAAAAAAATATAGTCTTGGAAAACAACCGACAGCTCATGGCAAAATTACAAAGTTTGAAACAAGCCAAGGCTGAACTAAGTGAAATCCTGAGGACTACATTAGAATGCAATTTAGTTGTCGAAAGAGATTTAACAAAGTTAAAATCAGATGTAGTAAAGCTCTACAATTTGAAGGATCGCCACAAAATGTGGTTGCAAGGGAGAGGGAAATCTGATGAATATTTAAAAGCTTTAGAGGAGAACAATATTCATAAATTAGATGAAATTAATGCTCATCGTGATATGGACACTTGGAAAGTTAATTGCACCCGGGAGACAGCTGAAAAACTGCTTGCCGGAAAACCACAGGGCACATTCCTGATAAGACCAAATTCTACAGGACAGTTAGCTCTCTCTATTGTCTGTAATAACATGGTCTATCATTGCATTATAAATAAGACTGAATGTGGATATGGTTTTGCTGAACCATACAATATCTACGAAACATTGAATGAATTAGTACTACATTATGCAGGAAATTCATTGGAGGAGCACAATGATCAGTTGAGAACTGAGTTAAAATACCCAGTTAATATGCATCTGGCAAAAAAACCTGAAAAGAATTGA
Protein
MVEIGVPQQSPEHSLEAAEWYWGDITRDEANEKLRDTRDGTFLVRNASNKDSGYTLTVRKGGANKLIKIYHHEGRYGFREPFEFKSVVDLVCYYSEYSLATCNSSLDIKLLYPLSRHQENEGGENISDETLATRYKEIHKKYILKTRDYNIRFSKYTRLREEVTLKRQALEAFREAVKMCEDHIKLQEKIQAEAQPHEKNIVLENNRQLMAKLQSLKQAKAELSEILRTTLECNLVVERDLTKLKSDVVKLYNLKDRHKMWLQGRGKSDEYLKALEENNIHKLDEINAHRDMDTWKVNCTRETAEKLLAGKPQGTFLIRPNSTGQLALSIVCNNMVYHCIINKTECGYGFAEPYNIYETLNELVLHYAGNSLEEHNDQLRTELKYPVNMHLAKKPEKN
Summary
Uniprot
B2MW85
E9JEI7
A0A2A4JYY8
A0A194PH65
A0A194R1T3
A0A212EPK6
+ More
A0A3S2TDL1 A0A0L7K582 A0A1B6CDR5 A0A2J7QUJ7 A0A1B6LVJ2 A0A0V0GB87 A0A069DX08 A0A023F1S3 A0A0H3W566 A0A1B6FJB3 A0A067QW74 A0A2R7WZ17 V9IIR3 E2B922 A0A0A9YVC8 K7IY25 A0A1L8DKY6 A0A310S655 A0A2A3EMQ0 A0A087ZTP6 A0A0L7RBK7 A0A026W141 A0A2A3ENF5 A0A232FBZ8 A0A0N0BDR0 E9ILF7 A0A3L8DWX7 A0A1Y1KG67 A0A1Y1KD31 A0A154PNQ7 A0A0J7NIY6 A0A195D5D4 F4X2C5 A0A2S2PZ81 E0VYA2 A0A158NJN6 A0A195BBL3 A0A195FNC5 E2ARR6 A0A195EKI9 T1J1N3 A0A151X6I9 J9JPF5 A0A1W5ALS3 A0A1W5AKH5 R7TP28 A0A2H8U1S2 A0A2S2P8K9 A0A1W5ADW3 A0A1A8CRB1 A0A1W4ZIJ2 B0S6V6 J9JXE8 A0A1W4ZHV8 A0A3B3QZV0 A0A1A8QBK6 A0A3Q3ERZ0 A0A3B3R0E5 A0A3Q1F2B0 A0A3B4XXY7 A0A3B4U8S0 A0A3Q2ZIJ4 A0A0T6B9V5 A0A3B3QX85 A0A3Q1CD09 A0A3P8RTX6 A0A3Q2Q6Y5 A0A2I4CPJ9 A0A1W4Z9B3 A0A1A7W9Z1 A0A2H6ND68 A0A3Q2C9S6 A0A3B5Q993 A0A091KBX4 A0A093BWZ0 A0A091LUJ8 A0A087VC22 A0A094KDT5 A0A091JWL0 A0A1S2ZRW3 A0A3B3R1D6 E1C8M4 A0A091RWK6 A0A2I0M1A2 A0A3B5M019 A0A1A8BEU9 A0A1A8KJT7 A0A2I4CPK2 A0A1U7RK29 A0A1W5A8R8 A0A3Q3K8C9 A0A3P8RC63
A0A3S2TDL1 A0A0L7K582 A0A1B6CDR5 A0A2J7QUJ7 A0A1B6LVJ2 A0A0V0GB87 A0A069DX08 A0A023F1S3 A0A0H3W566 A0A1B6FJB3 A0A067QW74 A0A2R7WZ17 V9IIR3 E2B922 A0A0A9YVC8 K7IY25 A0A1L8DKY6 A0A310S655 A0A2A3EMQ0 A0A087ZTP6 A0A0L7RBK7 A0A026W141 A0A2A3ENF5 A0A232FBZ8 A0A0N0BDR0 E9ILF7 A0A3L8DWX7 A0A1Y1KG67 A0A1Y1KD31 A0A154PNQ7 A0A0J7NIY6 A0A195D5D4 F4X2C5 A0A2S2PZ81 E0VYA2 A0A158NJN6 A0A195BBL3 A0A195FNC5 E2ARR6 A0A195EKI9 T1J1N3 A0A151X6I9 J9JPF5 A0A1W5ALS3 A0A1W5AKH5 R7TP28 A0A2H8U1S2 A0A2S2P8K9 A0A1W5ADW3 A0A1A8CRB1 A0A1W4ZIJ2 B0S6V6 J9JXE8 A0A1W4ZHV8 A0A3B3QZV0 A0A1A8QBK6 A0A3Q3ERZ0 A0A3B3R0E5 A0A3Q1F2B0 A0A3B4XXY7 A0A3B4U8S0 A0A3Q2ZIJ4 A0A0T6B9V5 A0A3B3QX85 A0A3Q1CD09 A0A3P8RTX6 A0A3Q2Q6Y5 A0A2I4CPJ9 A0A1W4Z9B3 A0A1A7W9Z1 A0A2H6ND68 A0A3Q2C9S6 A0A3B5Q993 A0A091KBX4 A0A093BWZ0 A0A091LUJ8 A0A087VC22 A0A094KDT5 A0A091JWL0 A0A1S2ZRW3 A0A3B3R1D6 E1C8M4 A0A091RWK6 A0A2I0M1A2 A0A3B5M019 A0A1A8BEU9 A0A1A8KJT7 A0A2I4CPK2 A0A1U7RK29 A0A1W5A8R8 A0A3Q3K8C9 A0A3P8RC63
Pubmed
EMBL
EU636066
ACC94001.1
BABH01005657
GQ426304
JQ087868
ADM32530.1
+ More
AEY80385.1 NWSH01000345 PCG77227.1 KQ459603 KPI92746.1 KQ460883 KPJ11210.1 AGBW02013442 OWR43430.1 RSAL01000288 RVE42936.1 JTDY01010846 KOB57957.1 GEDC01025878 GEDC01010233 JAS11420.1 JAS27065.1 NEVH01010552 PNF32256.1 GEBQ01012312 JAT27665.1 GECL01000753 JAP05371.1 GBGD01000469 JAC88420.1 GBBI01003250 JAC15462.1 KP635379 AKK23736.1 GECZ01019491 JAS50278.1 KK852886 KDR14361.1 KK856067 PTY24772.1 JR049394 AEY60950.1 GL446425 EFN87788.1 GBHO01010074 GDHC01021167 JAG33530.1 JAP97461.1 GFDF01006955 JAV07129.1 KQ768636 OAD53077.1 KZ288207 PBC33035.1 KQ414617 KOC68223.1 KK107499 EZA49782.1 PBC33034.1 NNAY01000442 OXU28354.1 KQ435851 KOX70804.1 GL764076 EFZ18596.1 QOIP01000003 RLU24802.1 GEZM01087900 JAV58406.1 GEZM01087899 JAV58408.1 KQ434977 KZC12898.1 LBMM01004387 KMQ92475.1 KQ976818 KYN08105.1 GL888575 EGI59407.1 GGMS01001538 MBY70741.1 DS235845 EEB18358.1 ADTU01018090 ADTU01018091 ADTU01018092 ADTU01018093 ADTU01018094 ADTU01018095 KQ976526 KYM81928.1 KQ981424 KYN41827.1 GL442166 EFN63874.1 KQ978782 KYN28384.1 JH431789 KQ982482 KYQ55939.1 ABLF02034706 AMQN01002403 KB309137 ELT95404.1 GFXV01008027 MBW19832.1 GGMR01013170 MBY25789.1 HADZ01018363 SBP82304.1 BX571667 ABLF02021400 HAEH01010832 SBR90717.1 LJIG01009142 KRT83671.1 HADW01000946 HADX01007476 SBP02346.1 IACI01084411 LAA29990.1 KK738976 KFP38064.1 KL449432 KFV04497.1 KK508204 KFP62127.1 KL487691 KFO10164.1 KL254687 KFZ56785.1 KK536441 KFP29552.1 AC186821 KK710105 KFQ32782.1 AKCR02000050 PKK23453.1 HADZ01001225 SBP65166.1 HAEE01012411 SBR32461.1
AEY80385.1 NWSH01000345 PCG77227.1 KQ459603 KPI92746.1 KQ460883 KPJ11210.1 AGBW02013442 OWR43430.1 RSAL01000288 RVE42936.1 JTDY01010846 KOB57957.1 GEDC01025878 GEDC01010233 JAS11420.1 JAS27065.1 NEVH01010552 PNF32256.1 GEBQ01012312 JAT27665.1 GECL01000753 JAP05371.1 GBGD01000469 JAC88420.1 GBBI01003250 JAC15462.1 KP635379 AKK23736.1 GECZ01019491 JAS50278.1 KK852886 KDR14361.1 KK856067 PTY24772.1 JR049394 AEY60950.1 GL446425 EFN87788.1 GBHO01010074 GDHC01021167 JAG33530.1 JAP97461.1 GFDF01006955 JAV07129.1 KQ768636 OAD53077.1 KZ288207 PBC33035.1 KQ414617 KOC68223.1 KK107499 EZA49782.1 PBC33034.1 NNAY01000442 OXU28354.1 KQ435851 KOX70804.1 GL764076 EFZ18596.1 QOIP01000003 RLU24802.1 GEZM01087900 JAV58406.1 GEZM01087899 JAV58408.1 KQ434977 KZC12898.1 LBMM01004387 KMQ92475.1 KQ976818 KYN08105.1 GL888575 EGI59407.1 GGMS01001538 MBY70741.1 DS235845 EEB18358.1 ADTU01018090 ADTU01018091 ADTU01018092 ADTU01018093 ADTU01018094 ADTU01018095 KQ976526 KYM81928.1 KQ981424 KYN41827.1 GL442166 EFN63874.1 KQ978782 KYN28384.1 JH431789 KQ982482 KYQ55939.1 ABLF02034706 AMQN01002403 KB309137 ELT95404.1 GFXV01008027 MBW19832.1 GGMR01013170 MBY25789.1 HADZ01018363 SBP82304.1 BX571667 ABLF02021400 HAEH01010832 SBR90717.1 LJIG01009142 KRT83671.1 HADW01000946 HADX01007476 SBP02346.1 IACI01084411 LAA29990.1 KK738976 KFP38064.1 KL449432 KFV04497.1 KK508204 KFP62127.1 KL487691 KFO10164.1 KL254687 KFZ56785.1 KK536441 KFP29552.1 AC186821 KK710105 KFQ32782.1 AKCR02000050 PKK23453.1 HADZ01001225 SBP65166.1 HAEE01012411 SBR32461.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000235965 UP000027135 UP000008237 UP000002358 UP000242457 UP000005203 UP000053825 UP000053097 UP000215335 UP000053105 UP000279307 UP000076502 UP000036403 UP000078542 UP000007755 UP000009046 UP000005205 UP000078540 UP000078541 UP000000311 UP000078492 UP000075809 UP000007819 UP000192224 UP000014760 UP000000437 UP000261540 UP000261660 UP000257200 UP000261360 UP000261420 UP000264800 UP000257160 UP000265080 UP000265000 UP000192220 UP000265020 UP000002852 UP000079721 UP000000539 UP000053872 UP000261380 UP000189705 UP000261600 UP000265100
UP000037510 UP000235965 UP000027135 UP000008237 UP000002358 UP000242457 UP000005203 UP000053825 UP000053097 UP000215335 UP000053105 UP000279307 UP000076502 UP000036403 UP000078542 UP000007755 UP000009046 UP000005205 UP000078540 UP000078541 UP000000311 UP000078492 UP000075809 UP000007819 UP000192224 UP000014760 UP000000437 UP000261540 UP000261660 UP000257200 UP000261360 UP000261420 UP000264800 UP000257160 UP000265080 UP000265000 UP000192220 UP000265020 UP000002852 UP000079721 UP000000539 UP000053872 UP000261380 UP000189705 UP000261600 UP000265100
Pfam
Interpro
IPR032498
PI3K_P85_iSH2
+ More
IPR036860 SH2_dom_sf
IPR000980 SH2
IPR035020 PI3kinase_P85_cSH2
IPR035022 PI3kinase_P85_nSH2
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001878 Znf_CCHC
IPR035979 RBD_domain_sf
IPR000198 RhoGAP_dom
IPR008936 Rho_GTPase_activation_prot
IPR001660 SAM
IPR013761 SAM/pointed_sf
IPR002219 PE/DAG-bd
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR036860 SH2_dom_sf
IPR000980 SH2
IPR035020 PI3kinase_P85_cSH2
IPR035022 PI3kinase_P85_nSH2
IPR000504 RRM_dom
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR001878 Znf_CCHC
IPR035979 RBD_domain_sf
IPR000198 RhoGAP_dom
IPR008936 Rho_GTPase_activation_prot
IPR001660 SAM
IPR013761 SAM/pointed_sf
IPR002219 PE/DAG-bd
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
B2MW85
E9JEI7
A0A2A4JYY8
A0A194PH65
A0A194R1T3
A0A212EPK6
+ More
A0A3S2TDL1 A0A0L7K582 A0A1B6CDR5 A0A2J7QUJ7 A0A1B6LVJ2 A0A0V0GB87 A0A069DX08 A0A023F1S3 A0A0H3W566 A0A1B6FJB3 A0A067QW74 A0A2R7WZ17 V9IIR3 E2B922 A0A0A9YVC8 K7IY25 A0A1L8DKY6 A0A310S655 A0A2A3EMQ0 A0A087ZTP6 A0A0L7RBK7 A0A026W141 A0A2A3ENF5 A0A232FBZ8 A0A0N0BDR0 E9ILF7 A0A3L8DWX7 A0A1Y1KG67 A0A1Y1KD31 A0A154PNQ7 A0A0J7NIY6 A0A195D5D4 F4X2C5 A0A2S2PZ81 E0VYA2 A0A158NJN6 A0A195BBL3 A0A195FNC5 E2ARR6 A0A195EKI9 T1J1N3 A0A151X6I9 J9JPF5 A0A1W5ALS3 A0A1W5AKH5 R7TP28 A0A2H8U1S2 A0A2S2P8K9 A0A1W5ADW3 A0A1A8CRB1 A0A1W4ZIJ2 B0S6V6 J9JXE8 A0A1W4ZHV8 A0A3B3QZV0 A0A1A8QBK6 A0A3Q3ERZ0 A0A3B3R0E5 A0A3Q1F2B0 A0A3B4XXY7 A0A3B4U8S0 A0A3Q2ZIJ4 A0A0T6B9V5 A0A3B3QX85 A0A3Q1CD09 A0A3P8RTX6 A0A3Q2Q6Y5 A0A2I4CPJ9 A0A1W4Z9B3 A0A1A7W9Z1 A0A2H6ND68 A0A3Q2C9S6 A0A3B5Q993 A0A091KBX4 A0A093BWZ0 A0A091LUJ8 A0A087VC22 A0A094KDT5 A0A091JWL0 A0A1S2ZRW3 A0A3B3R1D6 E1C8M4 A0A091RWK6 A0A2I0M1A2 A0A3B5M019 A0A1A8BEU9 A0A1A8KJT7 A0A2I4CPK2 A0A1U7RK29 A0A1W5A8R8 A0A3Q3K8C9 A0A3P8RC63
A0A3S2TDL1 A0A0L7K582 A0A1B6CDR5 A0A2J7QUJ7 A0A1B6LVJ2 A0A0V0GB87 A0A069DX08 A0A023F1S3 A0A0H3W566 A0A1B6FJB3 A0A067QW74 A0A2R7WZ17 V9IIR3 E2B922 A0A0A9YVC8 K7IY25 A0A1L8DKY6 A0A310S655 A0A2A3EMQ0 A0A087ZTP6 A0A0L7RBK7 A0A026W141 A0A2A3ENF5 A0A232FBZ8 A0A0N0BDR0 E9ILF7 A0A3L8DWX7 A0A1Y1KG67 A0A1Y1KD31 A0A154PNQ7 A0A0J7NIY6 A0A195D5D4 F4X2C5 A0A2S2PZ81 E0VYA2 A0A158NJN6 A0A195BBL3 A0A195FNC5 E2ARR6 A0A195EKI9 T1J1N3 A0A151X6I9 J9JPF5 A0A1W5ALS3 A0A1W5AKH5 R7TP28 A0A2H8U1S2 A0A2S2P8K9 A0A1W5ADW3 A0A1A8CRB1 A0A1W4ZIJ2 B0S6V6 J9JXE8 A0A1W4ZHV8 A0A3B3QZV0 A0A1A8QBK6 A0A3Q3ERZ0 A0A3B3R0E5 A0A3Q1F2B0 A0A3B4XXY7 A0A3B4U8S0 A0A3Q2ZIJ4 A0A0T6B9V5 A0A3B3QX85 A0A3Q1CD09 A0A3P8RTX6 A0A3Q2Q6Y5 A0A2I4CPJ9 A0A1W4Z9B3 A0A1A7W9Z1 A0A2H6ND68 A0A3Q2C9S6 A0A3B5Q993 A0A091KBX4 A0A093BWZ0 A0A091LUJ8 A0A087VC22 A0A094KDT5 A0A091JWL0 A0A1S2ZRW3 A0A3B3R1D6 E1C8M4 A0A091RWK6 A0A2I0M1A2 A0A3B5M019 A0A1A8BEU9 A0A1A8KJT7 A0A2I4CPK2 A0A1U7RK29 A0A1W5A8R8 A0A3Q3K8C9 A0A3P8RC63
PDB
5M6U
E-value=3.98703e-78,
Score=742
Ontologies
GO
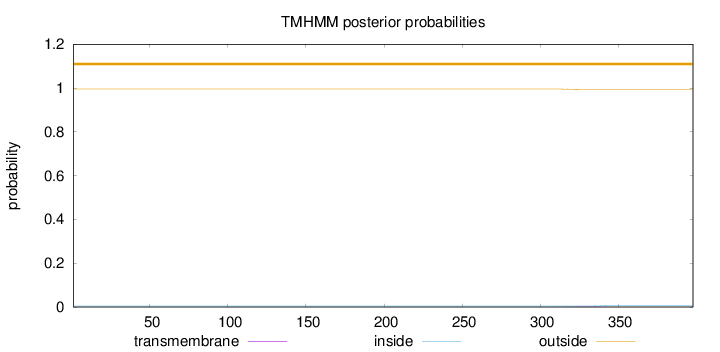
Topology
Length:
398
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03061
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00485
outside
1 - 398
Population Genetic Test Statistics
Pi
44.186658
Theta
177.797113
Tajima's D
-0.652461
CLR
1.083113
CSRT
0.206489675516224
Interpretation
Uncertain
