Gene
KWMTBOMO07174
Pre Gene Modal
BGIBMGA010397
Annotation
PREDICTED:_integral_membrane_protein_GPR180-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.865
Sequence
CDS
ATGTGTATTCAAGATCTCTTGATATTTTGTTGTCTTTTCGTTGTTGGATCATCGACGCACATAACAGGAGAGTTTTCTACTGCAGAATTCTTCAAGTTTTTAGTAAAATTTGGTTTCCAAAAGACCGACATACATTTTCAGAAAGAAACATATGGTTATATTTTTGGAAACATTACAGCAAACGATCACTTTAAACATCCCATAACATTTGCAGTTCTTGATAGAAGACATTTTTTACATTATTATAAGAATAGAGATATTATTGATAAAGAAATGGCATGTCAATCAATGTTCCAAAACTTAAATGGAACTGCCTACCATCCAAAATGTAATGCTTACGGTCAAGACCTATTTCGACGTGTACCTTGTATAAAGTCTGAACTATGTATTGACGAGGACACTCCATGGAATGTAATTAAGAAAAATCAATTTACCTACGTGATACAGAACACAGGGCAACCACGATTTTGGTATGTTTCTATGGTTGCCTGCTACTTAAATGAAGAAACTTGTTCATGGCATCACTATCCAGGAGCACCAAAAGACAACAAAACATTAAAAGCTATGCCACAAGCATTACAGTATGACTTCTGGTTAGTGAATGGAAGTCCAAACCTCTCATTCTACAATACATTGTTGTATCAATTTTCATTCGATCGACAAAATACATTGGAATTATATCTTATGTTTTGGCTATGTTATATTATATTGCTGCCCGTCCAGTTGTATGCAGTTAGAACACAAAAGCACCCGGTAACAAAACTGTTCACATTTAGTTTGGTCTTAGAGTTCATAGCTCTTTGTTTCAATGTTTTGCATACTGTCAAATTCGCTGTTGATGGAGTAGGATTTGCTGGTCTGTCAGTTGCAGGAGATATTCTTGATATTTTGAGTAGAACTTTGTTTATGTTACTACTCCTACTGTTGGCGAAAGGTTGGGCAGTGACGCGGTTGGAACTTACATACAAGCCCCTTGTTTTTGGAGTATGGATTATATATGGTGTGGTCCATATTTTATTGTACGTATGGAATACTATTGAGGTCGACATAATTGAAGACATCGATGAATACCAAACATGGCCGGGATGGTTGATTTTGGCTCTAAGGGTAGCCATCATGATATGGTTCGTCCTGGAATTACGTAACACGATGATGTACGAACATAATATGCCGAAATTGAACTTCTTTTTGCACTTTGGAGCTTCGAGCCTCGTATGGTATTACTTATTCGGCGGACTGTTTAGCTTTTTGTGTGATGGCGCATTTGTTATGGCCAACACGCTCAGAACAATACTTACTTCTTGCGCCTCCTAA
Protein
MCIQDLLIFCCLFVVGSSTHITGEFSTAEFFKFLVKFGFQKTDIHFQKETYGYIFGNITANDHFKHPITFAVLDRRHFLHYYKNRDIIDKEMACQSMFQNLNGTAYHPKCNAYGQDLFRRVPCIKSELCIDEDTPWNVIKKNQFTYVIQNTGQPRFWYVSMVACYLNEETCSWHHYPGAPKDNKTLKAMPQALQYDFWLVNGSPNLSFYNTLLYQFSFDRQNTLELYLMFWLCYIILLPVQLYAVRTQKHPVTKLFTFSLVLEFIALCFNVLHTVKFAVDGVGFAGLSVAGDILDILSRTLFMLLLLLLAKGWAVTRLELTYKPLVFGVWIIYGVVHILLYVWNTIEVDIIEDIDEYQTWPGWLILALRVAIMIWFVLELRNTMMYEHNMPKLNFFLHFGASSLVWYYLFGGLFSFLCDGAFVMANTLRTILTSCAS
Summary
Uniprot
A0A2A4K0S4
A0A1E1WKU2
A0A0L7KH00
A0A212EW85
A0A194PNP4
E9IYV6
+ More
A0A0J7NV49 A0A195F3W2 A0A0M8ZZD2 K7J5W3 A0A151XEI9 A0A195DBB5 F4WZ24 E2A0J4 A0A232EN22 A0A158NDM3 A0A195BQS1 A0A026VY08 H9JLJ6 E2C5E4 A0A088A7T3 A0A195CL80 A0A2A3ENP2 V5G6T3 A0A2P8ZB79 A0A0C9QKU7 A0A0L7RBA6 D6WF04 A0A0C9RWM6 A0A067R9D2 A0A1Y1MAY0 E0VRA0 T1HIB3 A0A146L419 A0A146LJ77 N6UGF3 J3JT91 A0A1B6E4V9 A0A0T6B5I7 A0A182VHS0 A0A182YJE7 A0A182PC68 A0A182R4C4 Q7PZ34 A0A182WEG5 Q16RT5 A0A182KCM8 A0A182L4F3 A0A182HH04 A0A182LXD2 A0A182N2J7 A0A084WBI5 B0WS88 A0A1Q3EWX8 A0A1A9UF76 A0A1A9YC70 A0A1B0BDL9 A0A182QWI8 A0A182IXH0 A0A1B0FNJ1 A0A2M3ZA19 A0A1Y1MAA6 A0A182FTB1 A0A2M4BID5 A0A2M4AQV4 W5JF78 B4KNP7 A0A0L0CM85 A0A1A9Z2D3 A0A1A9X3E5 A0A1I8MZ13 B4GHD1 B5E0N1 B4LM60 A0A0Q9WF86 A0A0A9XFT5 B3MJ26 A0A3B0J2K0 A0A1I8PCS2 A0A3B0J219 A0A0K8TF19 A0A1W4UZY3 A0A0M3QUN4 B4J6C2 A0A182XKP8 A0A194R5R8 Q8T056 B4MS39 A0A1L8D8W7 A0A1L8D8W8 B3NNA8 B4QGN0 Q9W2B3 A9UNI3 A0A1B0CRR7 A0A0J9RIM1 B4P8C8 A0A1J1ISN3 A0A336MGI7
A0A0J7NV49 A0A195F3W2 A0A0M8ZZD2 K7J5W3 A0A151XEI9 A0A195DBB5 F4WZ24 E2A0J4 A0A232EN22 A0A158NDM3 A0A195BQS1 A0A026VY08 H9JLJ6 E2C5E4 A0A088A7T3 A0A195CL80 A0A2A3ENP2 V5G6T3 A0A2P8ZB79 A0A0C9QKU7 A0A0L7RBA6 D6WF04 A0A0C9RWM6 A0A067R9D2 A0A1Y1MAY0 E0VRA0 T1HIB3 A0A146L419 A0A146LJ77 N6UGF3 J3JT91 A0A1B6E4V9 A0A0T6B5I7 A0A182VHS0 A0A182YJE7 A0A182PC68 A0A182R4C4 Q7PZ34 A0A182WEG5 Q16RT5 A0A182KCM8 A0A182L4F3 A0A182HH04 A0A182LXD2 A0A182N2J7 A0A084WBI5 B0WS88 A0A1Q3EWX8 A0A1A9UF76 A0A1A9YC70 A0A1B0BDL9 A0A182QWI8 A0A182IXH0 A0A1B0FNJ1 A0A2M3ZA19 A0A1Y1MAA6 A0A182FTB1 A0A2M4BID5 A0A2M4AQV4 W5JF78 B4KNP7 A0A0L0CM85 A0A1A9Z2D3 A0A1A9X3E5 A0A1I8MZ13 B4GHD1 B5E0N1 B4LM60 A0A0Q9WF86 A0A0A9XFT5 B3MJ26 A0A3B0J2K0 A0A1I8PCS2 A0A3B0J219 A0A0K8TF19 A0A1W4UZY3 A0A0M3QUN4 B4J6C2 A0A182XKP8 A0A194R5R8 Q8T056 B4MS39 A0A1L8D8W7 A0A1L8D8W8 B3NNA8 B4QGN0 Q9W2B3 A9UNI3 A0A1B0CRR7 A0A0J9RIM1 B4P8C8 A0A1J1ISN3 A0A336MGI7
Pubmed
26227816
22118469
26354079
21282665
20075255
21719571
+ More
20798317 28648823 21347285 24508170 30249741 19121390 29403074 18362917 19820115 24845553 28004739 20566863 26823975 23537049 22516182 25244985 12364791 17510324 20966253 24438588 20920257 23761445 17994087 26108605 25315136 15632085 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304
20798317 28648823 21347285 24508170 30249741 19121390 29403074 18362917 19820115 24845553 28004739 20566863 26823975 23537049 22516182 25244985 12364791 17510324 20966253 24438588 20920257 23761445 17994087 26108605 25315136 15632085 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304
EMBL
NWSH01000345
PCG77242.1
GDQN01003568
JAT87486.1
JTDY01009879
KOB62385.1
+ More
AGBW02012056 OWR45755.1 KQ459603 KPI92745.1 GL767051 EFZ14255.1 LBMM01001467 KMQ96270.1 KQ981820 KYN35148.1 KQ435793 KOX74210.1 KQ982254 KYQ58779.1 KQ981082 KYN09709.1 GL888465 EGI60527.1 GL435626 EFN72897.1 NNAY01003254 OXU19727.1 ADTU01012715 KQ976424 KYM88309.1 KK107578 QOIP01000002 EZA48678.1 RLU26100.1 BABH01005657 GL452770 EFN76877.1 KQ977600 KYN01481.1 KZ288203 PBC33385.1 GALX01002671 JAB65795.1 PYGN01000117 PSN53754.1 GBYB01004164 JAG73931.1 KQ414617 KOC68149.1 KQ971319 EFA00307.1 GBYB01012206 JAG81973.1 KK852846 KDR15115.1 GEZM01036366 JAV82761.1 DS235459 EEB15906.1 ACPB03006711 GDHC01015338 JAQ03291.1 GDHC01012129 JAQ06500.1 APGK01021647 APGK01021648 KB740350 KB632240 ENN80840.1 ERL90525.1 BT126447 AEE61411.1 GEDC01004337 JAS32961.1 LJIG01009650 KRT82656.1 AAAB01008986 EAA00082.3 CH477697 EAT37106.1 APCN01002370 AXCM01001386 ATLV01022359 KE525331 KFB47579.1 DS232066 EDS33715.1 GFDL01015224 JAV19821.1 JXJN01012518 AXCN02001076 CCAG010000365 GGFM01004622 MBW25373.1 GEZM01036365 JAV82762.1 GGFJ01003679 MBW52820.1 GGFK01009845 MBW43166.1 ADMH02001578 ETN61988.1 CH933808 EDW10032.2 JRES01000195 KNC33400.1 CH479183 EDW35901.1 CM000071 EDY69376.1 CH940648 EDW60938.2 KRF79678.1 GBHO01025971 JAG17633.1 CH902619 EDV37092.2 OUUW01000001 SPP75395.1 SPP75394.1 GBRD01001732 JAG64089.1 CP012524 ALC40969.1 CH916367 EDW00895.1 KQ460883 KPJ11211.1 AY069543 AAL39688.1 CH963850 EDW74928.1 GFDF01011182 JAV02902.1 GFDF01011181 JAV02903.1 CH954179 EDV55532.1 CM000362 EDX08149.1 AE013599 AAF46779.3 AHN56496.1 BT031347 ABY21760.1 AJWK01025161 AJWK01025162 AJWK01025163 AJWK01025164 CM002911 KMY95682.1 CM000158 EDW91168.2 CVRI01000059 CRL03243.1 UFQT01000928 SSX28039.1
AGBW02012056 OWR45755.1 KQ459603 KPI92745.1 GL767051 EFZ14255.1 LBMM01001467 KMQ96270.1 KQ981820 KYN35148.1 KQ435793 KOX74210.1 KQ982254 KYQ58779.1 KQ981082 KYN09709.1 GL888465 EGI60527.1 GL435626 EFN72897.1 NNAY01003254 OXU19727.1 ADTU01012715 KQ976424 KYM88309.1 KK107578 QOIP01000002 EZA48678.1 RLU26100.1 BABH01005657 GL452770 EFN76877.1 KQ977600 KYN01481.1 KZ288203 PBC33385.1 GALX01002671 JAB65795.1 PYGN01000117 PSN53754.1 GBYB01004164 JAG73931.1 KQ414617 KOC68149.1 KQ971319 EFA00307.1 GBYB01012206 JAG81973.1 KK852846 KDR15115.1 GEZM01036366 JAV82761.1 DS235459 EEB15906.1 ACPB03006711 GDHC01015338 JAQ03291.1 GDHC01012129 JAQ06500.1 APGK01021647 APGK01021648 KB740350 KB632240 ENN80840.1 ERL90525.1 BT126447 AEE61411.1 GEDC01004337 JAS32961.1 LJIG01009650 KRT82656.1 AAAB01008986 EAA00082.3 CH477697 EAT37106.1 APCN01002370 AXCM01001386 ATLV01022359 KE525331 KFB47579.1 DS232066 EDS33715.1 GFDL01015224 JAV19821.1 JXJN01012518 AXCN02001076 CCAG010000365 GGFM01004622 MBW25373.1 GEZM01036365 JAV82762.1 GGFJ01003679 MBW52820.1 GGFK01009845 MBW43166.1 ADMH02001578 ETN61988.1 CH933808 EDW10032.2 JRES01000195 KNC33400.1 CH479183 EDW35901.1 CM000071 EDY69376.1 CH940648 EDW60938.2 KRF79678.1 GBHO01025971 JAG17633.1 CH902619 EDV37092.2 OUUW01000001 SPP75395.1 SPP75394.1 GBRD01001732 JAG64089.1 CP012524 ALC40969.1 CH916367 EDW00895.1 KQ460883 KPJ11211.1 AY069543 AAL39688.1 CH963850 EDW74928.1 GFDF01011182 JAV02902.1 GFDF01011181 JAV02903.1 CH954179 EDV55532.1 CM000362 EDX08149.1 AE013599 AAF46779.3 AHN56496.1 BT031347 ABY21760.1 AJWK01025161 AJWK01025162 AJWK01025163 AJWK01025164 CM002911 KMY95682.1 CM000158 EDW91168.2 CVRI01000059 CRL03243.1 UFQT01000928 SSX28039.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053268
UP000036403
UP000078541
+ More
UP000053105 UP000002358 UP000075809 UP000078492 UP000007755 UP000000311 UP000215335 UP000005205 UP000078540 UP000053097 UP000279307 UP000005204 UP000008237 UP000005203 UP000078542 UP000242457 UP000245037 UP000053825 UP000007266 UP000027135 UP000009046 UP000015103 UP000019118 UP000030742 UP000075903 UP000076408 UP000075885 UP000075900 UP000007062 UP000075920 UP000008820 UP000075881 UP000075882 UP000075840 UP000075883 UP000075884 UP000030765 UP000002320 UP000078200 UP000092443 UP000092460 UP000075886 UP000075880 UP000092444 UP000069272 UP000000673 UP000009192 UP000037069 UP000092445 UP000091820 UP000095301 UP000008744 UP000001819 UP000008792 UP000007801 UP000268350 UP000095300 UP000192221 UP000092553 UP000001070 UP000076407 UP000053240 UP000007798 UP000008711 UP000000304 UP000000803 UP000092461 UP000002282 UP000183832
UP000053105 UP000002358 UP000075809 UP000078492 UP000007755 UP000000311 UP000215335 UP000005205 UP000078540 UP000053097 UP000279307 UP000005204 UP000008237 UP000005203 UP000078542 UP000242457 UP000245037 UP000053825 UP000007266 UP000027135 UP000009046 UP000015103 UP000019118 UP000030742 UP000075903 UP000076408 UP000075885 UP000075900 UP000007062 UP000075920 UP000008820 UP000075881 UP000075882 UP000075840 UP000075883 UP000075884 UP000030765 UP000002320 UP000078200 UP000092443 UP000092460 UP000075886 UP000075880 UP000092444 UP000069272 UP000000673 UP000009192 UP000037069 UP000092445 UP000091820 UP000095301 UP000008744 UP000001819 UP000008792 UP000007801 UP000268350 UP000095300 UP000192221 UP000092553 UP000001070 UP000076407 UP000053240 UP000007798 UP000008711 UP000000304 UP000000803 UP000092461 UP000002282 UP000183832
Pfam
PF10192 GpcrRhopsn4
Interpro
IPR019336
Intimal_thickness-rel_rcpt
ProteinModelPortal
A0A2A4K0S4
A0A1E1WKU2
A0A0L7KH00
A0A212EW85
A0A194PNP4
E9IYV6
+ More
A0A0J7NV49 A0A195F3W2 A0A0M8ZZD2 K7J5W3 A0A151XEI9 A0A195DBB5 F4WZ24 E2A0J4 A0A232EN22 A0A158NDM3 A0A195BQS1 A0A026VY08 H9JLJ6 E2C5E4 A0A088A7T3 A0A195CL80 A0A2A3ENP2 V5G6T3 A0A2P8ZB79 A0A0C9QKU7 A0A0L7RBA6 D6WF04 A0A0C9RWM6 A0A067R9D2 A0A1Y1MAY0 E0VRA0 T1HIB3 A0A146L419 A0A146LJ77 N6UGF3 J3JT91 A0A1B6E4V9 A0A0T6B5I7 A0A182VHS0 A0A182YJE7 A0A182PC68 A0A182R4C4 Q7PZ34 A0A182WEG5 Q16RT5 A0A182KCM8 A0A182L4F3 A0A182HH04 A0A182LXD2 A0A182N2J7 A0A084WBI5 B0WS88 A0A1Q3EWX8 A0A1A9UF76 A0A1A9YC70 A0A1B0BDL9 A0A182QWI8 A0A182IXH0 A0A1B0FNJ1 A0A2M3ZA19 A0A1Y1MAA6 A0A182FTB1 A0A2M4BID5 A0A2M4AQV4 W5JF78 B4KNP7 A0A0L0CM85 A0A1A9Z2D3 A0A1A9X3E5 A0A1I8MZ13 B4GHD1 B5E0N1 B4LM60 A0A0Q9WF86 A0A0A9XFT5 B3MJ26 A0A3B0J2K0 A0A1I8PCS2 A0A3B0J219 A0A0K8TF19 A0A1W4UZY3 A0A0M3QUN4 B4J6C2 A0A182XKP8 A0A194R5R8 Q8T056 B4MS39 A0A1L8D8W7 A0A1L8D8W8 B3NNA8 B4QGN0 Q9W2B3 A9UNI3 A0A1B0CRR7 A0A0J9RIM1 B4P8C8 A0A1J1ISN3 A0A336MGI7
A0A0J7NV49 A0A195F3W2 A0A0M8ZZD2 K7J5W3 A0A151XEI9 A0A195DBB5 F4WZ24 E2A0J4 A0A232EN22 A0A158NDM3 A0A195BQS1 A0A026VY08 H9JLJ6 E2C5E4 A0A088A7T3 A0A195CL80 A0A2A3ENP2 V5G6T3 A0A2P8ZB79 A0A0C9QKU7 A0A0L7RBA6 D6WF04 A0A0C9RWM6 A0A067R9D2 A0A1Y1MAY0 E0VRA0 T1HIB3 A0A146L419 A0A146LJ77 N6UGF3 J3JT91 A0A1B6E4V9 A0A0T6B5I7 A0A182VHS0 A0A182YJE7 A0A182PC68 A0A182R4C4 Q7PZ34 A0A182WEG5 Q16RT5 A0A182KCM8 A0A182L4F3 A0A182HH04 A0A182LXD2 A0A182N2J7 A0A084WBI5 B0WS88 A0A1Q3EWX8 A0A1A9UF76 A0A1A9YC70 A0A1B0BDL9 A0A182QWI8 A0A182IXH0 A0A1B0FNJ1 A0A2M3ZA19 A0A1Y1MAA6 A0A182FTB1 A0A2M4BID5 A0A2M4AQV4 W5JF78 B4KNP7 A0A0L0CM85 A0A1A9Z2D3 A0A1A9X3E5 A0A1I8MZ13 B4GHD1 B5E0N1 B4LM60 A0A0Q9WF86 A0A0A9XFT5 B3MJ26 A0A3B0J2K0 A0A1I8PCS2 A0A3B0J219 A0A0K8TF19 A0A1W4UZY3 A0A0M3QUN4 B4J6C2 A0A182XKP8 A0A194R5R8 Q8T056 B4MS39 A0A1L8D8W7 A0A1L8D8W8 B3NNA8 B4QGN0 Q9W2B3 A9UNI3 A0A1B0CRR7 A0A0J9RIM1 B4P8C8 A0A1J1ISN3 A0A336MGI7
Ontologies
GO
Topology
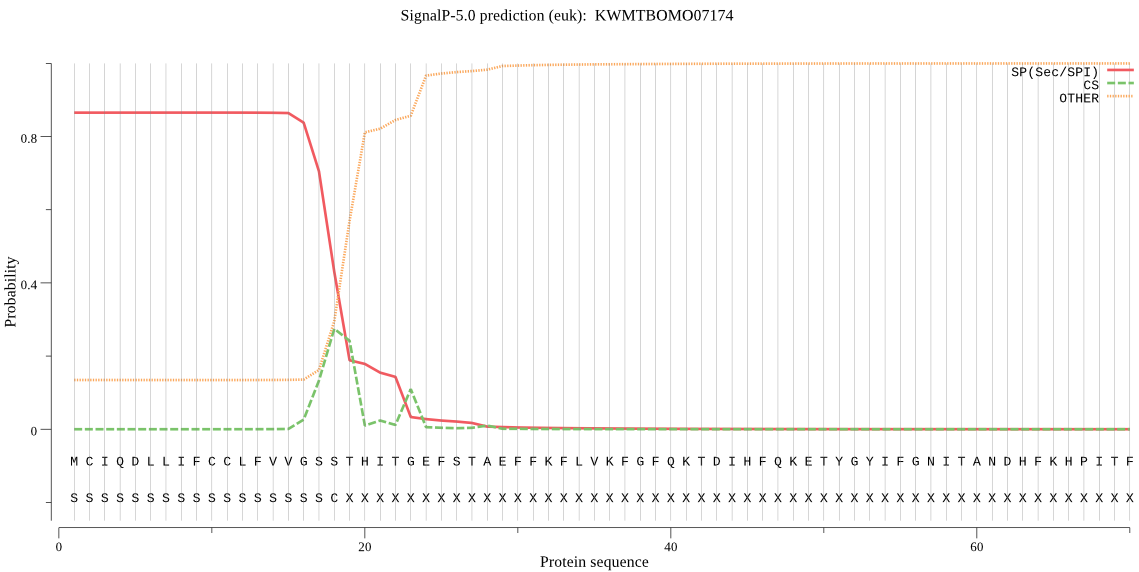
SignalP
Position: 1 - 18,
Likelihood: 0.865433
Length:
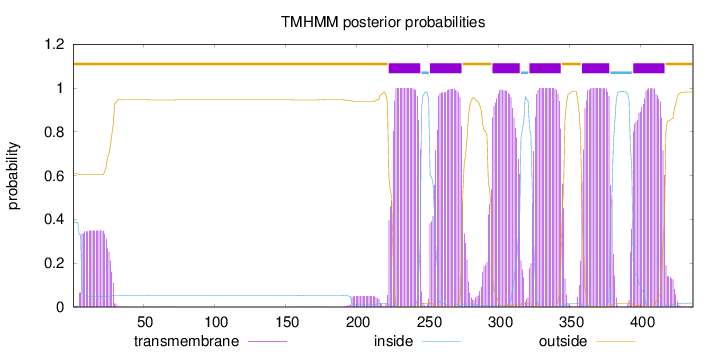
437
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
136.75656
Exp number, first 60 AAs:
7.37603
Total prob of N-in:
0.39058
outside
1 - 222
TMhelix
223 - 245
inside
246 - 251
TMhelix
252 - 274
outside
275 - 295
TMhelix
296 - 315
inside
316 - 321
TMhelix
322 - 344
outside
345 - 358
TMhelix
359 - 378
inside
379 - 394
TMhelix
395 - 417
outside
418 - 437
Population Genetic Test Statistics
Pi
166.434696
Theta
164.919143
Tajima's D
0.21307
CLR
0.02653
CSRT
0.432328383580821
Interpretation
Uncertain
