Gene
KWMTBOMO07172
Pre Gene Modal
BGIBMGA010560
Annotation
PREDICTED:_protein_retinal_degeneration_B_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.83
Sequence
CDS
ATGCTTATTAAAGAATACAGAATACCGCTACCACTCACGGTTGAAGAATACAGAATAGCACAACTTTATATGATTGCAAAGAAAAGTAGGGAAGAAAGCTCTGGTGAAGGAAGTGGTGTAGAAATTTTAGTCAACGAACCATATGAAGATGGGCCAGGTGGCAAAGGTCAGTACACTCAAAAAATATACCATGTTGGTAGCCATTTACCAGGTTGGTTTAAGAGTCTCCTCCCAAAATCTGCATTAACAGTATCTGAGGAAGCTTGGAATGCTTATCCTTACACAAAAACTAGGTACACATGTCCATTTGTTGAAAAATTTTCACTGGAAATTGAAACTTACTATTATGCTGACAATGGCCACCAAGAAAATGTATTTGACCTTTCTGGGAGTGACTTAAAGAACAGAATAGTTGATGTTATTGATGTTGTTAAAGATCAACTGTATGGTACTGATTACATAAAGGAAGAGGATCCAAAATTATTTATTTCACAGAAGTGTAATCGAGGTCCTTTGTCTGATAACTGGTTAGAAGAGTATTGGACAGAGGTACAAGGACAGCCTCAGCCATTAGCTAATGGAAAATCTCTCATGTGTGCCTATAAATTATGTAGAGTAGAATTCAGGTATTGGGGCATGCAAACAAAATTGGAAAAATTTATTCATGATGTTGCATTAAGGAAAACTATGTTGCGAGCTCATAGACAAGCCTGGGCTTGGCAAGATGAATGGCATGGTCTAACAATGGATGATATTAGAGAAATTGAAAGACAGACCCAATTGGCTCTACAGAAAAAAATGTGTGGTGATTCACAGGGTGAGGAACAAGATGTATCAGATGAAAATTCTAAATCCCTTGCAGCTACAATGAGTAGCTTAGAAAAAAATGAAGAATTGACAACCCCTTTGACAAACAAGAAAAATCATAATGAAAAACGCCTTCAAAAACATTTAAGTCCAGAGATTACTCCTCCTTCAGATCGTAAATCAACATCAAAGTCGAATCTCAGATCTTCATCCTCTGGCTCAATAAAAAGTTTGCAGGCTCAAGCTGCAAACTGGCGAATGGAAACACTTGTCAGAGAATCAGAAACAGAAACGGGGTCAGAAGATGAATTTTATGATTGTGAATCTTCTTTTAACAAATGGTCTTCTATGTGCTCTCTTGATGAAGCTGATATTGATATTTCACCCATTCAAGGTGATCTAGGAACTCAAGATAGCATTTTTAATCCTTCTTTCTTAAAACGTGTTACTTCTGAGAGAGGATCTCGGAGAGTAGTAACTTTACAAAGCCACCACAGTGTTGATGGATGTCCTGACACACCAGAACATGGCTCATGCGCTACTACCGTATTGATACTAGTATTTCATGCAGGAAGTGTACTAGACGCAAATGTTGACATGACAGCTAAAAAATCTGATGTTACTACCTTTAAAGGAGCTTTTGAATCAGTAATGCGACAACATTATCCAACTTTAGTTGGACATGTTATTATAAAATTAGTATCTTGTCCTTCCATATGCACTGAATCACTTGGAATATTATCAACATTAAGCCCTTATAGTTTTGATTGTTCTCCATCTACTGTTGAAACACCATCACTAACTAATGATCTTATACCTATTGGTGCTATACCTCTAATAGCCACAACTTGTCCCGAATATCATGAGTCTGTTACAAAAACCATTAACTCAGCTAATGCTGTATTTAACGAGTTTATTAAATCTGAGGATGGAAAAGGTTTCAATGGTCAAGTTTGTATTGTGGGCGATAGTATGGGGTCTGTGCTGGCTTACGACGCACTTGTTAGAAGTTTCCAGTATAACTCACGACACGATAGCGAAAATAGTATATTAGATACTGAAATTACGATTCCAAATGATCCCTTAGATCAACAATATCTAAACAAATCTCATTTACAAGCGCCTACGCCGCGAAGGCGATCGTCTTCTACCAGCGAAAATCAAATAAAATTCGAATTCGAAGTCAGTGATTTTTTTACTTTTGGTAGTCCATTATCATTGATTTTAGCTTCTAGGAAAGTATCAGATGACAGAAGTCGAGATATTACAAAACCACCGGTTCAGCAGGTTTACAATTTATTTCACCCAACTGATCCAGTTGCTTCAAGATTAGAACCATTACTATCAGCAAGATTCACAAATTTGCCCCCCATAAATGTTGCTAGATATACGAAGTATCCATTAGGAAATGGTCAGCCTTATCATTTGATGGAATTAATACAAAGTCATCCTCAATTGTTTGGTGACCATTTACAAATGCCACCTACACCAATTTTACGTAGACTCTCTGAAGCGTCTGTACAAAGCACAGTTAGTGGTTTAGTTGATAATATTCCTTTGTTAACTATGAATGCTTTACAGCAGAAATGGTGGGGTTCTAAACGCTTAGATTACGCCTTATATTGTCCTGAAGGCTTAACCAACTTCCCCACGAATGCTCTTCCTCATCTATTTCATGCAAGTTACTGGGAAAGTTACGACGTTATAGCTTTCATATTACGACAGATCGGTCATTTTGATTTAGCTTTGTATGGACATTCAGATGATAAGGACTGCTCTATGTTTAAACCAGGCCAAGAAAGAGAAAAATGGATGAGAAAGAGAACATCAGTTAAATTAAAAAATGTCGCCGCAAATCATAGAGCCAATGATGTAATAGTGAAAGAAGGTTCTTCGCAAGTTTTCGTTGCTCGTTTTATGTATGGACCTTTGGACATGATAACTCTAACAGGAGAAAAGGTTGATATATATATGATGAAAGACCCACCAGCAGGTGAATGGACACTTTTATCAACTGTCGTGACAGATAAAACGGGAAGAATCTCTTATACATTACCGGAGCGTCAAAGTGTTGGTTGTGGTATTTATCCTGTCAAAGCGGTTGTTAGAGGTGATCACACTAGTTGTAATTTTCACTTAGCTGTTGTTCCACCTCAAACCGAATGCGTTGTTTTTAGTATTGACGGTTCTTTTACCGCTAGCGTTTCTGTAACTGGACGCGACCCAAAAGTAAGAGCTGGTGCAGTTGATGTTGTTCGTTTTTGGCAAGATTTGGGCTATCTCATTCTGTATATTACTGGTAGACCTGATATGCAACAAAGGAAAGTAATATCCTGGTTAGCAGAACACAACTTCCCCCATGGCTTAGTCTTTTTCTCGGACGGATTTTCAACAGACCCCTTGGGACATAAGGCCGCACATTTGAACGGACTTATTAACGATCATGGCATAATTCTACATGCTGCGTACGGTTCGGGAAAAGACATAACGGTCTATAACAATTGTGGTTTGTCTCCTAGACAAATTTATGCCATTGGTCGCATTGGCAAGAAATACAGCAACATGGCTACTACTTTAAGTGATGGATACGCTTCACATTTGGCAGATTTGAAACAGCCTGGGGCAATTCGCCCTGCTAGAGGTAATGCCCGTCTTCTCGTCCCTCGTCGATTGTTAGCTCCGGTTACAAATGCACCTGCTTCCCGTGGTCGCCGTTAG
Protein
MLIKEYRIPLPLTVEEYRIAQLYMIAKKSREESSGEGSGVEILVNEPYEDGPGGKGQYTQKIYHVGSHLPGWFKSLLPKSALTVSEEAWNAYPYTKTRYTCPFVEKFSLEIETYYYADNGHQENVFDLSGSDLKNRIVDVIDVVKDQLYGTDYIKEEDPKLFISQKCNRGPLSDNWLEEYWTEVQGQPQPLANGKSLMCAYKLCRVEFRYWGMQTKLEKFIHDVALRKTMLRAHRQAWAWQDEWHGLTMDDIREIERQTQLALQKKMCGDSQGEEQDVSDENSKSLAATMSSLEKNEELTTPLTNKKNHNEKRLQKHLSPEITPPSDRKSTSKSNLRSSSSGSIKSLQAQAANWRMETLVRESETETGSEDEFYDCESSFNKWSSMCSLDEADIDISPIQGDLGTQDSIFNPSFLKRVTSERGSRRVVTLQSHHSVDGCPDTPEHGSCATTVLILVFHAGSVLDANVDMTAKKSDVTTFKGAFESVMRQHYPTLVGHVIIKLVSCPSICTESLGILSTLSPYSFDCSPSTVETPSLTNDLIPIGAIPLIATTCPEYHESVTKTINSANAVFNEFIKSEDGKGFNGQVCIVGDSMGSVLAYDALVRSFQYNSRHDSENSILDTEITIPNDPLDQQYLNKSHLQAPTPRRRSSSTSENQIKFEFEVSDFFTFGSPLSLILASRKVSDDRSRDITKPPVQQVYNLFHPTDPVASRLEPLLSARFTNLPPINVARYTKYPLGNGQPYHLMELIQSHPQLFGDHLQMPPTPILRRLSEASVQSTVSGLVDNIPLLTMNALQQKWWGSKRLDYALYCPEGLTNFPTNALPHLFHASYWESYDVIAFILRQIGHFDLALYGHSDDKDCSMFKPGQEREKWMRKRTSVKLKNVAANHRANDVIVKEGSSQVFVARFMYGPLDMITLTGEKVDIYMMKDPPAGEWTLLSTVVTDKTGRISYTLPERQSVGCGIYPVKAVVRGDHTSCNFHLAVVPPQTECVVFSIDGSFTASVSVTGRDPKVRAGAVDVVRFWQDLGYLILYITGRPDMQQRKVISWLAEHNFPHGLVFFSDGFSTDPLGHKAAHLNGLINDHGIILHAAYGSGKDITVYNNCGLSPRQIYAIGRIGKKYSNMATTLSDGYASHLADLKQPGAIRPARGNARLLVPRRLLAPVTNAPASRGRR
Summary
Uniprot
H9JM08
A0A2A4K0N7
A0A0L7LBZ2
A0A212FLT0
A0A194PJ46
A0A194R0I4
+ More
D6WG59 A0A139WLJ0 A0A067QZE4 A0A310SJ13 A0A0C9PMW4 A0A0C9R3F1 A0A232F2J4 A0A0C9RMX5 A0A026VV22 K7IPU2 A0A2A3EPM4 E2BBQ0 A0A088AVI9 E2A1Y9 V9I8U5 V9I982 A0A0M8ZPJ4 A0A158P207 A0A151IKY0 F4W681 A0A1J1IQG0 U4UI36 A0A195AW75 A0A0A9VS85 A0A0A9VVJ9 A0A154PN32 N6T4I8 A0A1L8DZ25 A0A0L7RJ54 A0A0K8WDP3 A0A0K8U5K9 A0A0K8V6L6 W8AXQ0 A0A0K8VYE9 A0A0K8UZX4 A0A0K8VVW1 A0A0A1X8S4 A0A0K8VRN0 A0A0K8W890 A0A034VD76 W8ALJ2 A0A1A9VSW0 A0A1B0GFR4 A0A0A1X5Q1 A0A0A1XDC1 A0A023F4I0 A0A1I8P371 A0A1I8P313 A0A1B0B6J2 A0A0L0BKY7 A0A1B0A1S6 A0A146M0E8 A0A034VB35 W8B8R8 A0A1I8N9Q2 A0A0A1WIH7 A0A1I8N9Q3 A0A1I8N9P8 A0A1I8N9P5 A0A0M4EZ44 T1IFQ0 A0A1A9YMA0 A0A1B6HB32 E0VPA5 A0A226DIM2 A0A2S2R9V3 J9K2G4 A0A0V0G8K5 A0A2H4WAM8 A0A1B6EH36 V5I8U2 A0A0F7VHP7 A0A3S3RY36 B7PQR9 A0A0K8W8D9 A0A1Y1NA70 A0A1Y1N5F8 R7V5K0 A0A2S2QEI6 A0A3B3VDG1 W5LJZ4 A0A3Q2DJU9 A0A1Q3G3V6 A0A1Y1N929 A0A3Q3MKY6 F1RFJ7 A0A1Y1N5J5 A0A3Q4BP59
D6WG59 A0A139WLJ0 A0A067QZE4 A0A310SJ13 A0A0C9PMW4 A0A0C9R3F1 A0A232F2J4 A0A0C9RMX5 A0A026VV22 K7IPU2 A0A2A3EPM4 E2BBQ0 A0A088AVI9 E2A1Y9 V9I8U5 V9I982 A0A0M8ZPJ4 A0A158P207 A0A151IKY0 F4W681 A0A1J1IQG0 U4UI36 A0A195AW75 A0A0A9VS85 A0A0A9VVJ9 A0A154PN32 N6T4I8 A0A1L8DZ25 A0A0L7RJ54 A0A0K8WDP3 A0A0K8U5K9 A0A0K8V6L6 W8AXQ0 A0A0K8VYE9 A0A0K8UZX4 A0A0K8VVW1 A0A0A1X8S4 A0A0K8VRN0 A0A0K8W890 A0A034VD76 W8ALJ2 A0A1A9VSW0 A0A1B0GFR4 A0A0A1X5Q1 A0A0A1XDC1 A0A023F4I0 A0A1I8P371 A0A1I8P313 A0A1B0B6J2 A0A0L0BKY7 A0A1B0A1S6 A0A146M0E8 A0A034VB35 W8B8R8 A0A1I8N9Q2 A0A0A1WIH7 A0A1I8N9Q3 A0A1I8N9P8 A0A1I8N9P5 A0A0M4EZ44 T1IFQ0 A0A1A9YMA0 A0A1B6HB32 E0VPA5 A0A226DIM2 A0A2S2R9V3 J9K2G4 A0A0V0G8K5 A0A2H4WAM8 A0A1B6EH36 V5I8U2 A0A0F7VHP7 A0A3S3RY36 B7PQR9 A0A0K8W8D9 A0A1Y1NA70 A0A1Y1N5F8 R7V5K0 A0A2S2QEI6 A0A3B3VDG1 W5LJZ4 A0A3Q2DJU9 A0A1Q3G3V6 A0A1Y1N929 A0A3Q3MKY6 F1RFJ7 A0A1Y1N5J5 A0A3Q4BP59
Pubmed
EMBL
BABH01005653
NWSH01000345
PCG77240.1
JTDY01001777
KOB72915.1
AGBW02007731
+ More
OWR54650.1 KQ459603 KPI92744.1 KQ460883 KPJ11212.1 KQ971320 EFA00532.2 KYB28802.1 KK853076 KDR11738.1 KQ759817 OAD62778.1 GBYB01002498 JAG72265.1 GBYB01002495 GBYB01002496 JAG72262.1 JAG72263.1 NNAY01001125 OXU25046.1 GBYB01014762 JAG84529.1 KK107837 EZA47530.1 KZ288198 PBC33667.1 GL447151 EFN86826.1 GL435870 EFN72549.1 JR036064 AEY56901.1 JR036061 JR036062 AEY56898.1 AEY56899.1 KQ435922 KOX68465.1 ADTU01006929 KQ977199 KYN05007.1 GL887707 EGI70216.1 CVRI01000056 CRL01780.1 KB632366 ERL93644.1 KQ976731 KYM76285.1 GBHO01044387 JAF99216.1 GBHO01044383 JAF99220.1 KQ434998 KZC13276.1 APGK01052899 APGK01052900 KB741216 ENN72528.1 GFDF01002371 JAV11713.1 KQ414582 KOC70885.1 GDHF01003048 JAI49266.1 GDHF01030441 JAI21873.1 GDHF01017798 JAI34516.1 GAMC01016982 JAB89573.1 GDHF01008448 JAI43866.1 GDHF01020105 JAI32209.1 GDHF01009303 JAI43011.1 GBXI01007214 JAD07078.1 GDHF01010816 JAI41498.1 GDHF01004963 JAI47351.1 GAKP01019255 GAKP01019252 JAC39700.1 GAMC01016980 JAB89575.1 CCAG010014042 GBXI01007855 JAD06437.1 GBXI01005337 JAD08955.1 GBBI01002630 JAC16082.1 JXJN01009116 JRES01001705 KNC20731.1 GDHC01005730 JAQ12899.1 GAKP01019253 JAC39699.1 GAMC01016984 JAB89571.1 GBXI01015468 JAC98823.1 CP012528 ALC49367.1 ACPB03005597 ACPB03005598 ACPB03005599 GECU01035839 JAS71867.1 DS235361 EEB15211.1 LNIX01000020 OXA44036.1 GGMS01017618 MBY86821.1 ABLF02030848 ABLF02030858 ABLF02059694 GECL01001822 JAP04302.1 MG020534 AUC64090.1 GEDC01028556 GEDC01017286 GEDC01000059 JAS08742.1 JAS20012.1 JAS37239.1 GALX01004090 JAB64376.1 LN830735 CFW94244.1 NCKU01003324 RWS07747.1 ABJB010061859 ABJB010101686 ABJB010143426 ABJB010148606 ABJB010175315 ABJB010316315 ABJB010403582 ABJB010498213 ABJB010717668 ABJB011110390 DS766923 EEC08941.1 GDHF01004846 JAI47468.1 GEZM01012188 GEZM01012187 JAV93126.1 GEZM01012191 JAV93123.1 AMQN01000748 KB295062 ELU13727.1 GGMS01006727 MBY75930.1 GFDL01000557 JAV34488.1 GEZM01012189 GEZM01012186 JAV93125.1 AEMK02000096 DQIR01101632 HDA57108.1 GEZM01012190 JAV93124.1
OWR54650.1 KQ459603 KPI92744.1 KQ460883 KPJ11212.1 KQ971320 EFA00532.2 KYB28802.1 KK853076 KDR11738.1 KQ759817 OAD62778.1 GBYB01002498 JAG72265.1 GBYB01002495 GBYB01002496 JAG72262.1 JAG72263.1 NNAY01001125 OXU25046.1 GBYB01014762 JAG84529.1 KK107837 EZA47530.1 KZ288198 PBC33667.1 GL447151 EFN86826.1 GL435870 EFN72549.1 JR036064 AEY56901.1 JR036061 JR036062 AEY56898.1 AEY56899.1 KQ435922 KOX68465.1 ADTU01006929 KQ977199 KYN05007.1 GL887707 EGI70216.1 CVRI01000056 CRL01780.1 KB632366 ERL93644.1 KQ976731 KYM76285.1 GBHO01044387 JAF99216.1 GBHO01044383 JAF99220.1 KQ434998 KZC13276.1 APGK01052899 APGK01052900 KB741216 ENN72528.1 GFDF01002371 JAV11713.1 KQ414582 KOC70885.1 GDHF01003048 JAI49266.1 GDHF01030441 JAI21873.1 GDHF01017798 JAI34516.1 GAMC01016982 JAB89573.1 GDHF01008448 JAI43866.1 GDHF01020105 JAI32209.1 GDHF01009303 JAI43011.1 GBXI01007214 JAD07078.1 GDHF01010816 JAI41498.1 GDHF01004963 JAI47351.1 GAKP01019255 GAKP01019252 JAC39700.1 GAMC01016980 JAB89575.1 CCAG010014042 GBXI01007855 JAD06437.1 GBXI01005337 JAD08955.1 GBBI01002630 JAC16082.1 JXJN01009116 JRES01001705 KNC20731.1 GDHC01005730 JAQ12899.1 GAKP01019253 JAC39699.1 GAMC01016984 JAB89571.1 GBXI01015468 JAC98823.1 CP012528 ALC49367.1 ACPB03005597 ACPB03005598 ACPB03005599 GECU01035839 JAS71867.1 DS235361 EEB15211.1 LNIX01000020 OXA44036.1 GGMS01017618 MBY86821.1 ABLF02030848 ABLF02030858 ABLF02059694 GECL01001822 JAP04302.1 MG020534 AUC64090.1 GEDC01028556 GEDC01017286 GEDC01000059 JAS08742.1 JAS20012.1 JAS37239.1 GALX01004090 JAB64376.1 LN830735 CFW94244.1 NCKU01003324 RWS07747.1 ABJB010061859 ABJB010101686 ABJB010143426 ABJB010148606 ABJB010175315 ABJB010316315 ABJB010403582 ABJB010498213 ABJB010717668 ABJB011110390 DS766923 EEC08941.1 GDHF01004846 JAI47468.1 GEZM01012188 GEZM01012187 JAV93126.1 GEZM01012191 JAV93123.1 AMQN01000748 KB295062 ELU13727.1 GGMS01006727 MBY75930.1 GFDL01000557 JAV34488.1 GEZM01012189 GEZM01012186 JAV93125.1 AEMK02000096 DQIR01101632 HDA57108.1 GEZM01012190 JAV93124.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000007266 UP000027135 UP000215335 UP000053097 UP000002358 UP000242457 UP000008237 UP000005203 UP000000311 UP000053105 UP000005205 UP000078542 UP000007755 UP000183832 UP000030742 UP000078540 UP000076502 UP000019118 UP000053825 UP000078200 UP000092444 UP000095300 UP000092460 UP000037069 UP000092445 UP000095301 UP000092553 UP000015103 UP000092443 UP000009046 UP000198287 UP000007819 UP000285301 UP000001555 UP000014760 UP000261500 UP000018467 UP000265020 UP000261640 UP000008227 UP000261620
UP000007266 UP000027135 UP000215335 UP000053097 UP000002358 UP000242457 UP000008237 UP000005203 UP000000311 UP000053105 UP000005205 UP000078542 UP000007755 UP000183832 UP000030742 UP000078540 UP000076502 UP000019118 UP000053825 UP000078200 UP000092444 UP000095300 UP000092460 UP000037069 UP000092445 UP000095301 UP000092553 UP000015103 UP000092443 UP000009046 UP000198287 UP000007819 UP000285301 UP000001555 UP000014760 UP000261500 UP000018467 UP000265020 UP000261640 UP000008227 UP000261620
PRIDE
Pfam
Interpro
IPR004177
DDHD_dom
+ More
IPR023393 START-like_dom_sf
IPR036412 HAD-like_sf
IPR031315 LNS2/PITP
IPR001666 PI_transfer
IPR023214 HAD_sf
IPR028158 RPA_interact_N_dom
IPR028159 RPA_interact_C_dom
IPR019140 MCM_complex-bd
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR007110 Ig-like_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR023393 START-like_dom_sf
IPR036412 HAD-like_sf
IPR031315 LNS2/PITP
IPR001666 PI_transfer
IPR023214 HAD_sf
IPR028158 RPA_interact_N_dom
IPR028159 RPA_interact_C_dom
IPR019140 MCM_complex-bd
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR007110 Ig-like_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
Gene 3D
CDD
ProteinModelPortal
H9JM08
A0A2A4K0N7
A0A0L7LBZ2
A0A212FLT0
A0A194PJ46
A0A194R0I4
+ More
D6WG59 A0A139WLJ0 A0A067QZE4 A0A310SJ13 A0A0C9PMW4 A0A0C9R3F1 A0A232F2J4 A0A0C9RMX5 A0A026VV22 K7IPU2 A0A2A3EPM4 E2BBQ0 A0A088AVI9 E2A1Y9 V9I8U5 V9I982 A0A0M8ZPJ4 A0A158P207 A0A151IKY0 F4W681 A0A1J1IQG0 U4UI36 A0A195AW75 A0A0A9VS85 A0A0A9VVJ9 A0A154PN32 N6T4I8 A0A1L8DZ25 A0A0L7RJ54 A0A0K8WDP3 A0A0K8U5K9 A0A0K8V6L6 W8AXQ0 A0A0K8VYE9 A0A0K8UZX4 A0A0K8VVW1 A0A0A1X8S4 A0A0K8VRN0 A0A0K8W890 A0A034VD76 W8ALJ2 A0A1A9VSW0 A0A1B0GFR4 A0A0A1X5Q1 A0A0A1XDC1 A0A023F4I0 A0A1I8P371 A0A1I8P313 A0A1B0B6J2 A0A0L0BKY7 A0A1B0A1S6 A0A146M0E8 A0A034VB35 W8B8R8 A0A1I8N9Q2 A0A0A1WIH7 A0A1I8N9Q3 A0A1I8N9P8 A0A1I8N9P5 A0A0M4EZ44 T1IFQ0 A0A1A9YMA0 A0A1B6HB32 E0VPA5 A0A226DIM2 A0A2S2R9V3 J9K2G4 A0A0V0G8K5 A0A2H4WAM8 A0A1B6EH36 V5I8U2 A0A0F7VHP7 A0A3S3RY36 B7PQR9 A0A0K8W8D9 A0A1Y1NA70 A0A1Y1N5F8 R7V5K0 A0A2S2QEI6 A0A3B3VDG1 W5LJZ4 A0A3Q2DJU9 A0A1Q3G3V6 A0A1Y1N929 A0A3Q3MKY6 F1RFJ7 A0A1Y1N5J5 A0A3Q4BP59
D6WG59 A0A139WLJ0 A0A067QZE4 A0A310SJ13 A0A0C9PMW4 A0A0C9R3F1 A0A232F2J4 A0A0C9RMX5 A0A026VV22 K7IPU2 A0A2A3EPM4 E2BBQ0 A0A088AVI9 E2A1Y9 V9I8U5 V9I982 A0A0M8ZPJ4 A0A158P207 A0A151IKY0 F4W681 A0A1J1IQG0 U4UI36 A0A195AW75 A0A0A9VS85 A0A0A9VVJ9 A0A154PN32 N6T4I8 A0A1L8DZ25 A0A0L7RJ54 A0A0K8WDP3 A0A0K8U5K9 A0A0K8V6L6 W8AXQ0 A0A0K8VYE9 A0A0K8UZX4 A0A0K8VVW1 A0A0A1X8S4 A0A0K8VRN0 A0A0K8W890 A0A034VD76 W8ALJ2 A0A1A9VSW0 A0A1B0GFR4 A0A0A1X5Q1 A0A0A1XDC1 A0A023F4I0 A0A1I8P371 A0A1I8P313 A0A1B0B6J2 A0A0L0BKY7 A0A1B0A1S6 A0A146M0E8 A0A034VB35 W8B8R8 A0A1I8N9Q2 A0A0A1WIH7 A0A1I8N9Q3 A0A1I8N9P8 A0A1I8N9P5 A0A0M4EZ44 T1IFQ0 A0A1A9YMA0 A0A1B6HB32 E0VPA5 A0A226DIM2 A0A2S2R9V3 J9K2G4 A0A0V0G8K5 A0A2H4WAM8 A0A1B6EH36 V5I8U2 A0A0F7VHP7 A0A3S3RY36 B7PQR9 A0A0K8W8D9 A0A1Y1NA70 A0A1Y1N5F8 R7V5K0 A0A2S2QEI6 A0A3B3VDG1 W5LJZ4 A0A3Q2DJU9 A0A1Q3G3V6 A0A1Y1N929 A0A3Q3MKY6 F1RFJ7 A0A1Y1N5J5 A0A3Q4BP59
PDB
1T27
E-value=1.32034e-54,
Score=543
Ontologies
GO
PANTHER
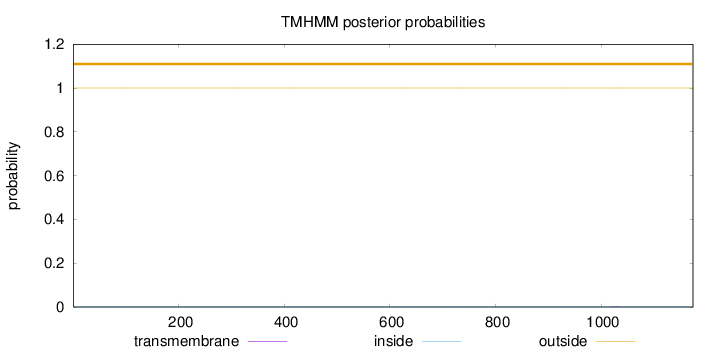
Topology
Length:
1174
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00678999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1174
Population Genetic Test Statistics
Pi
199.931841
Theta
182.254591
Tajima's D
-0.643926
CLR
0.527119
CSRT
0.212939353032348
Interpretation
Uncertain
