Gene
KWMTBOMO07171 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010559
Annotation
PREDICTED:_carnitine_O-palmitoyltransferase_2?_mitochondrial_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.662
Sequence
CDS
ATGTTATCTTTGAGAGAACATATTTTGGAACCAGAAGTTTATCATTTGAATGCGAAAAAGAGTGATACTCCGTTATTTAGAACAGTTACTGGATTATTACCTGAAGCTATCTCTTGGTATGGTGCATATTTATTCAAAGCATTTCCATTAGATATGAGTCAATTTGTTGGACTATTTGGAGCTACACGTTTACCTCAAATAGGTAAAGATAAATTATTCAGGGATCCTAAGAGCAAGCATGTTTTAGTCCAAAGAAGGGGAAGATTTTATGCATTTGATGTCTTAGATAAGGACGGCAACCTATTGTCTCCTGCTGAATTACTTGGAAACCTATCTAAAATTATTAGAGATGACACCGGTTCGGCTGAACATCCCCTAGGTTACTTGACCTGCCAAACTAGAGATTTGTGGGCGAAACAAAGAGCTCACCTTGAAGAAAATGGAAATAAACAAGCATTACATGTATTAGATACAGCTATATTCAACTTGATATTTGACGATGATGTTATAGAAGATGATAAAAATAAACTTCTGAAACATTATTTGTGTGGTAATGGAAATAACAGGTGGTTTGATAAATCATTCAGCTTAATTGTTACAAAGGATGGAGTGTCTGGGATAAATTTTGAACACTCTTGGGGAGATGGAGTAGCTGTGCTAAGGTTCTTCCAAGACATATACAATGAAACAACAACAAAACCTTTTGTACATCCAGATTCTCAACCATCAACCTCAAACATTACTGTTCAAGAACTACAATTCAACCTTGATGAAAAATCGAAGAAGTTTATTCAGGACGCAAAGCAAGAATATGATAACTGGTGTGACTCCTTGAGCATTGATTACATATTATATGAAGGCCTCACCAAAGGAGCTTGCAAGAAATTCAAAGTTAGCCCAGATTGTATAATGCAACTAGCTTTTCAAGCGGCATATCATCTGATAAAGGGTAAATTCGTTGGAACCTACGAGTCTTGCAGCACCTCTGCCTTTAAACATGGACGTACTGAAACAATGCGACCGTGCACCGAAAAAACTAAGGCATTCTGCGAAGCTTTAAATGTTCATAAAAAGTCGAAAGATGAACTTCGAGTAGCTATGGGCGAGTGTTCCGCCTATCACTCTGAGTTAGTAAAACAAGCTGCCATGGGCCAAGGCTTCGATAGACACTTGTTCGCTCTTATGAAGATCGCTGAAGAAAATGATATACCAAGACCAGAATTATTCGATTCATACGAATATAAGTTCCTAAACAAATCAATTCTTAGTACAAGTACTTTGTCGGCTCCCAGTGTAATGGCAGGTGGATTTGGACCAGTTGCACAGGAGGGATTCGGTATTGGGTACTCGGCTTTTCCTGATAAACTAGGTGCAGCTGTGAGCAGTTATAAAACGCACAATAGTAGTACCGAATTTGTGCAAGCTCTTCATACATCGTTTTTAGAAATCACAAGGATTTTGTCTGGATAG
Protein
MLSLREHILEPEVYHLNAKKSDTPLFRTVTGLLPEAISWYGAYLFKAFPLDMSQFVGLFGATRLPQIGKDKLFRDPKSKHVLVQRRGRFYAFDVLDKDGNLLSPAELLGNLSKIIRDDTGSAEHPLGYLTCQTRDLWAKQRAHLEENGNKQALHVLDTAIFNLIFDDDVIEDDKNKLLKHYLCGNGNNRWFDKSFSLIVTKDGVSGINFEHSWGDGVAVLRFFQDIYNETTTKPFVHPDSQPSTSNITVQELQFNLDEKSKKFIQDAKQEYDNWCDSLSIDYILYEGLTKGACKKFKVSPDCIMQLAFQAAYHLIKGKFVGTYESCSTSAFKHGRTETMRPCTEKTKAFCEALNVHKKSKDELRVAMGECSAYHSELVKQAAMGQGFDRHLFALMKIAEENDIPRPELFDSYEYKFLNKSILSTSTLSAPSVMAGGFGPVAQEGFGIGYSAFPDKLGAAVSSYKTHNSSTEFVQALHTSFLEITRILSG
Summary
Similarity
Belongs to the carnitine/choline acetyltransferase family.
Uniprot
H9JM07
A0A194R095
A0A194PI71
A0A291P163
A0A0P0ELC5
A0A1V0M8B1
+ More
A0A291P115 A0A212FLS5 A0A088MD10 A0A3S2LRC0 A0A2A4JZK2 A0A2H1V5I8 A0A182MZ77 A0A182QK97 A0A182P8A9 A0A1Q3F3U5 A0A182JI10 A0A195CZQ6 A0A195EFA6 A0A182JUM5 A0A182M7Q6 A0A158NRT9 E9IY55 A0A195ERN7 F4WQR7 Q5TNJ3 A0A195BPY1 A0A182XNP7 A0A182GMS2 A0A151X3T5 A0A182LMU8 A0A182UI32 B0WRQ6 K7ITB3 A0A182I7A6 A0A2J7RSV9 A0A182VUD5 A0A182USB4 A0A026WCT2 A0A0J7L7M2 A0A182Y0B1 A0A0P4VNB7 Q171R6 U5ESD7 A0A0V0G6I2 A0A2J7RSV8 A0A023F332 A0A067R1Z9 A0A069DW47 A0A2M4BH49 A0A2M4AKL0 A0A0P6FS08 A0A2M3ZA45 A0A224XLP3 A0A0P4Y2B2 A0A0P5D6U6 A0A0P5CUQ9 A0A0N7ZVN9 A0A0P5F8U5 A0A154PGT5 A0A0P5WWA4 A0A0P5A4B6 A0A0P5FHS2 A0A0P5VC46 A0A182F2K0 E2AEF2 A0A0N7ZQQ0 A0A0K8TTB4 E9FY65 A0A232F9D9 A0A0N8C9K2 A0A1B6IBM3 W5JW02 V5G906 A0A0P5L5K3 A0A0P5VSP2 A0A0P5R985 N6U5K5 A0A1B6CFU1 A0A2R2MQ02 A0A0L7RB32 A0A093H524 A0A0K8THC3 A0A0C9RAR1 A0A1U9W5K6 A0A091EQP7 B4LE41 A0A0A9W8I4 R0JG34 A0A088A7X0 A0A1Y1MAA5 A0A093NWE3 A0A336MKE1 A0A2A3EP17 U3INM7 A0A226NAI1 A0A087RHS9 A0A226PRE9 J9JKJ9 A0A0P5IPE7
A0A291P115 A0A212FLS5 A0A088MD10 A0A3S2LRC0 A0A2A4JZK2 A0A2H1V5I8 A0A182MZ77 A0A182QK97 A0A182P8A9 A0A1Q3F3U5 A0A182JI10 A0A195CZQ6 A0A195EFA6 A0A182JUM5 A0A182M7Q6 A0A158NRT9 E9IY55 A0A195ERN7 F4WQR7 Q5TNJ3 A0A195BPY1 A0A182XNP7 A0A182GMS2 A0A151X3T5 A0A182LMU8 A0A182UI32 B0WRQ6 K7ITB3 A0A182I7A6 A0A2J7RSV9 A0A182VUD5 A0A182USB4 A0A026WCT2 A0A0J7L7M2 A0A182Y0B1 A0A0P4VNB7 Q171R6 U5ESD7 A0A0V0G6I2 A0A2J7RSV8 A0A023F332 A0A067R1Z9 A0A069DW47 A0A2M4BH49 A0A2M4AKL0 A0A0P6FS08 A0A2M3ZA45 A0A224XLP3 A0A0P4Y2B2 A0A0P5D6U6 A0A0P5CUQ9 A0A0N7ZVN9 A0A0P5F8U5 A0A154PGT5 A0A0P5WWA4 A0A0P5A4B6 A0A0P5FHS2 A0A0P5VC46 A0A182F2K0 E2AEF2 A0A0N7ZQQ0 A0A0K8TTB4 E9FY65 A0A232F9D9 A0A0N8C9K2 A0A1B6IBM3 W5JW02 V5G906 A0A0P5L5K3 A0A0P5VSP2 A0A0P5R985 N6U5K5 A0A1B6CFU1 A0A2R2MQ02 A0A0L7RB32 A0A093H524 A0A0K8THC3 A0A0C9RAR1 A0A1U9W5K6 A0A091EQP7 B4LE41 A0A0A9W8I4 R0JG34 A0A088A7X0 A0A1Y1MAA5 A0A093NWE3 A0A336MKE1 A0A2A3EP17 U3INM7 A0A226NAI1 A0A087RHS9 A0A226PRE9 J9JKJ9 A0A0P5IPE7
Pubmed
19121390
26354079
28986331
26445454
28349297
22118469
+ More
26385554 21347285 21282665 21719571 12364791 14747013 17210077 26483478 20966253 20075255 24508170 30249741 25244985 27129103 17510324 25474469 24845553 26334808 20798317 26369729 21292972 28648823 20920257 23761445 23537049 17994087 25401762 28004739 24621616
26385554 21347285 21282665 21719571 12364791 14747013 17210077 26483478 20966253 20075255 24508170 30249741 25244985 27129103 17510324 25474469 24845553 26334808 20798317 26369729 21292972 28648823 20920257 23761445 23537049 17994087 25401762 28004739 24621616
EMBL
BABH01005653
KQ460883
KPJ11213.1
KQ459603
KPI92743.1
MF706177
+ More
ATJ44604.1 KT261721 ALJ30261.1 KU755509 ARD71219.1 MF687650 ATJ44576.1 AGBW02007731 OWR54649.1 KJ579232 AIN34708.1 RSAL01000299 RVE42779.1 NWSH01000345 PCG77239.1 ODYU01000563 SOQ35534.1 AXCN02002069 GFDL01012820 JAV22225.1 KQ977041 KYN06143.1 KQ978983 KYN26841.1 AXCM01000345 ADTU01024307 GL766908 EFZ14502.1 KQ981993 KYN30888.1 GL888275 EGI63430.1 AAAB01008984 EAL38971.2 KQ976424 KYM88577.1 JXUM01075043 KQ562865 KXJ74976.1 KQ982562 KYQ54939.1 DS232058 EDS33460.1 AAZX01005891 APCN01003374 NEVH01000250 PNF43916.1 KK107274 QOIP01000008 EZA53748.1 RLU19713.1 LBMM01000373 KMQ98991.1 GDKW01002176 JAI54419.1 CH477447 EAT40751.1 GANO01002403 JAB57468.1 GECL01003090 JAP03034.1 PNF43912.1 GBBI01003064 JAC15648.1 KK852808 KDR16023.1 GBGD01000823 JAC88066.1 GGFJ01002987 MBW52128.1 GGFK01007999 MBW41320.1 GDIQ01046258 JAN48479.1 GGFM01004652 MBW25403.1 GFTR01007487 JAW08939.1 GDIP01237334 JAI86067.1 GDIP01164639 LRGB01002371 JAJ58763.1 KZS08048.1 GDIP01165268 JAJ58134.1 GDIP01208025 JAJ15377.1 GDIP01150387 JAJ73015.1 KQ434899 KZC11017.1 GDIP01081584 GDIP01070453 JAM22131.1 GDIP01204282 JAJ19120.1 GDIQ01254391 JAJ97333.1 GDIP01101729 JAM01986.1 GL438828 EFN68204.1 GDIP01221937 JAJ01465.1 GDAI01000205 JAI17398.1 GL732527 EFX87483.1 NNAY01000676 OXU27053.1 GDIQ01101443 JAL50283.1 GECU01023372 JAS84334.1 ADMH02000239 ETN67280.1 GALX01001842 GALX01001841 JAB66625.1 GDIQ01174656 JAK77069.1 GDIP01096942 JAM06773.1 GDIQ01110780 JAL40946.1 APGK01039173 KB740967 ENN76890.1 GEDC01025093 JAS12205.1 KQ414617 KOC68074.1 KL217220 KFV74520.1 GBRD01001286 JAG64535.1 GBYB01013559 GBYB01013560 JAG83326.1 JAG83327.1 KX892708 AQY19419.1 KK718817 KFO58614.1 CH940647 EDW70084.1 GBHO01038833 GBHO01038832 GBHO01038830 GBHO01038829 JAG04771.1 JAG04772.1 JAG04774.1 JAG04775.1 KB744139 EOA95911.1 GEZM01037797 JAV81928.1 KL224816 KFW64447.1 UFQS01001396 UFQT01001396 SSX10673.1 SSX30355.1 KZ288203 PBC33440.1 MCFN01000116 OXB64594.1 KL226373 KFM13033.1 AWGT02000029 OXB82090.1 ABLF02016211 ABLF02016213 GDIQ01209838 JAK41887.1
ATJ44604.1 KT261721 ALJ30261.1 KU755509 ARD71219.1 MF687650 ATJ44576.1 AGBW02007731 OWR54649.1 KJ579232 AIN34708.1 RSAL01000299 RVE42779.1 NWSH01000345 PCG77239.1 ODYU01000563 SOQ35534.1 AXCN02002069 GFDL01012820 JAV22225.1 KQ977041 KYN06143.1 KQ978983 KYN26841.1 AXCM01000345 ADTU01024307 GL766908 EFZ14502.1 KQ981993 KYN30888.1 GL888275 EGI63430.1 AAAB01008984 EAL38971.2 KQ976424 KYM88577.1 JXUM01075043 KQ562865 KXJ74976.1 KQ982562 KYQ54939.1 DS232058 EDS33460.1 AAZX01005891 APCN01003374 NEVH01000250 PNF43916.1 KK107274 QOIP01000008 EZA53748.1 RLU19713.1 LBMM01000373 KMQ98991.1 GDKW01002176 JAI54419.1 CH477447 EAT40751.1 GANO01002403 JAB57468.1 GECL01003090 JAP03034.1 PNF43912.1 GBBI01003064 JAC15648.1 KK852808 KDR16023.1 GBGD01000823 JAC88066.1 GGFJ01002987 MBW52128.1 GGFK01007999 MBW41320.1 GDIQ01046258 JAN48479.1 GGFM01004652 MBW25403.1 GFTR01007487 JAW08939.1 GDIP01237334 JAI86067.1 GDIP01164639 LRGB01002371 JAJ58763.1 KZS08048.1 GDIP01165268 JAJ58134.1 GDIP01208025 JAJ15377.1 GDIP01150387 JAJ73015.1 KQ434899 KZC11017.1 GDIP01081584 GDIP01070453 JAM22131.1 GDIP01204282 JAJ19120.1 GDIQ01254391 JAJ97333.1 GDIP01101729 JAM01986.1 GL438828 EFN68204.1 GDIP01221937 JAJ01465.1 GDAI01000205 JAI17398.1 GL732527 EFX87483.1 NNAY01000676 OXU27053.1 GDIQ01101443 JAL50283.1 GECU01023372 JAS84334.1 ADMH02000239 ETN67280.1 GALX01001842 GALX01001841 JAB66625.1 GDIQ01174656 JAK77069.1 GDIP01096942 JAM06773.1 GDIQ01110780 JAL40946.1 APGK01039173 KB740967 ENN76890.1 GEDC01025093 JAS12205.1 KQ414617 KOC68074.1 KL217220 KFV74520.1 GBRD01001286 JAG64535.1 GBYB01013559 GBYB01013560 JAG83326.1 JAG83327.1 KX892708 AQY19419.1 KK718817 KFO58614.1 CH940647 EDW70084.1 GBHO01038833 GBHO01038832 GBHO01038830 GBHO01038829 JAG04771.1 JAG04772.1 JAG04774.1 JAG04775.1 KB744139 EOA95911.1 GEZM01037797 JAV81928.1 KL224816 KFW64447.1 UFQS01001396 UFQT01001396 SSX10673.1 SSX30355.1 KZ288203 PBC33440.1 MCFN01000116 OXB64594.1 KL226373 KFM13033.1 AWGT02000029 OXB82090.1 ABLF02016211 ABLF02016213 GDIQ01209838 JAK41887.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000283053
UP000218220
+ More
UP000075884 UP000075886 UP000075885 UP000075880 UP000078542 UP000078492 UP000075881 UP000075883 UP000005205 UP000078541 UP000007755 UP000007062 UP000078540 UP000076407 UP000069940 UP000249989 UP000075809 UP000075882 UP000075902 UP000002320 UP000002358 UP000075840 UP000235965 UP000075920 UP000075903 UP000053097 UP000279307 UP000036403 UP000076408 UP000008820 UP000027135 UP000076858 UP000076502 UP000069272 UP000000311 UP000000305 UP000215335 UP000000673 UP000019118 UP000085678 UP000053825 UP000053875 UP000052976 UP000008792 UP000005203 UP000054081 UP000242457 UP000198323 UP000053286 UP000198419 UP000007819
UP000075884 UP000075886 UP000075885 UP000075880 UP000078542 UP000078492 UP000075881 UP000075883 UP000005205 UP000078541 UP000007755 UP000007062 UP000078540 UP000076407 UP000069940 UP000249989 UP000075809 UP000075882 UP000075902 UP000002320 UP000002358 UP000075840 UP000235965 UP000075920 UP000075903 UP000053097 UP000279307 UP000036403 UP000076408 UP000008820 UP000027135 UP000076858 UP000076502 UP000069272 UP000000311 UP000000305 UP000215335 UP000000673 UP000019118 UP000085678 UP000053825 UP000053875 UP000052976 UP000008792 UP000005203 UP000054081 UP000242457 UP000198323 UP000053286 UP000198419 UP000007819
PRIDE
Interpro
ProteinModelPortal
H9JM07
A0A194R095
A0A194PI71
A0A291P163
A0A0P0ELC5
A0A1V0M8B1
+ More
A0A291P115 A0A212FLS5 A0A088MD10 A0A3S2LRC0 A0A2A4JZK2 A0A2H1V5I8 A0A182MZ77 A0A182QK97 A0A182P8A9 A0A1Q3F3U5 A0A182JI10 A0A195CZQ6 A0A195EFA6 A0A182JUM5 A0A182M7Q6 A0A158NRT9 E9IY55 A0A195ERN7 F4WQR7 Q5TNJ3 A0A195BPY1 A0A182XNP7 A0A182GMS2 A0A151X3T5 A0A182LMU8 A0A182UI32 B0WRQ6 K7ITB3 A0A182I7A6 A0A2J7RSV9 A0A182VUD5 A0A182USB4 A0A026WCT2 A0A0J7L7M2 A0A182Y0B1 A0A0P4VNB7 Q171R6 U5ESD7 A0A0V0G6I2 A0A2J7RSV8 A0A023F332 A0A067R1Z9 A0A069DW47 A0A2M4BH49 A0A2M4AKL0 A0A0P6FS08 A0A2M3ZA45 A0A224XLP3 A0A0P4Y2B2 A0A0P5D6U6 A0A0P5CUQ9 A0A0N7ZVN9 A0A0P5F8U5 A0A154PGT5 A0A0P5WWA4 A0A0P5A4B6 A0A0P5FHS2 A0A0P5VC46 A0A182F2K0 E2AEF2 A0A0N7ZQQ0 A0A0K8TTB4 E9FY65 A0A232F9D9 A0A0N8C9K2 A0A1B6IBM3 W5JW02 V5G906 A0A0P5L5K3 A0A0P5VSP2 A0A0P5R985 N6U5K5 A0A1B6CFU1 A0A2R2MQ02 A0A0L7RB32 A0A093H524 A0A0K8THC3 A0A0C9RAR1 A0A1U9W5K6 A0A091EQP7 B4LE41 A0A0A9W8I4 R0JG34 A0A088A7X0 A0A1Y1MAA5 A0A093NWE3 A0A336MKE1 A0A2A3EP17 U3INM7 A0A226NAI1 A0A087RHS9 A0A226PRE9 J9JKJ9 A0A0P5IPE7
A0A291P115 A0A212FLS5 A0A088MD10 A0A3S2LRC0 A0A2A4JZK2 A0A2H1V5I8 A0A182MZ77 A0A182QK97 A0A182P8A9 A0A1Q3F3U5 A0A182JI10 A0A195CZQ6 A0A195EFA6 A0A182JUM5 A0A182M7Q6 A0A158NRT9 E9IY55 A0A195ERN7 F4WQR7 Q5TNJ3 A0A195BPY1 A0A182XNP7 A0A182GMS2 A0A151X3T5 A0A182LMU8 A0A182UI32 B0WRQ6 K7ITB3 A0A182I7A6 A0A2J7RSV9 A0A182VUD5 A0A182USB4 A0A026WCT2 A0A0J7L7M2 A0A182Y0B1 A0A0P4VNB7 Q171R6 U5ESD7 A0A0V0G6I2 A0A2J7RSV8 A0A023F332 A0A067R1Z9 A0A069DW47 A0A2M4BH49 A0A2M4AKL0 A0A0P6FS08 A0A2M3ZA45 A0A224XLP3 A0A0P4Y2B2 A0A0P5D6U6 A0A0P5CUQ9 A0A0N7ZVN9 A0A0P5F8U5 A0A154PGT5 A0A0P5WWA4 A0A0P5A4B6 A0A0P5FHS2 A0A0P5VC46 A0A182F2K0 E2AEF2 A0A0N7ZQQ0 A0A0K8TTB4 E9FY65 A0A232F9D9 A0A0N8C9K2 A0A1B6IBM3 W5JW02 V5G906 A0A0P5L5K3 A0A0P5VSP2 A0A0P5R985 N6U5K5 A0A1B6CFU1 A0A2R2MQ02 A0A0L7RB32 A0A093H524 A0A0K8THC3 A0A0C9RAR1 A0A1U9W5K6 A0A091EQP7 B4LE41 A0A0A9W8I4 R0JG34 A0A088A7X0 A0A1Y1MAA5 A0A093NWE3 A0A336MKE1 A0A2A3EP17 U3INM7 A0A226NAI1 A0A087RHS9 A0A226PRE9 J9JKJ9 A0A0P5IPE7
PDB
4EPH
E-value=9.17303e-127,
Score=1162
Ontologies
PATHWAY
GO
PANTHER
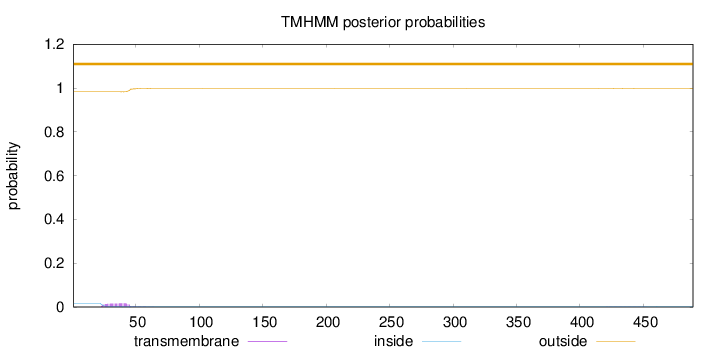
Topology
Length:
489
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.36922
Exp number, first 60 AAs:
0.35583
Total prob of N-in:
0.01694
outside
1 - 489
Population Genetic Test Statistics
Pi
153.469424
Theta
126.858328
Tajima's D
-0.621571
CLR
137.885426
CSRT
0.210289485525724
Interpretation
Uncertain
