Gene
KWMTBOMO07170
Pre Gene Modal
BGIBMGA010398
Annotation
PREDICTED:_4F2_cell-surface_antigen_heavy_chain-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.825
Sequence
CDS
ATGGACAGCTTACCAGAGAACAGTACTAATGGTGGAAGTCAGTTAGAGACTTCTGATTATTTGCATGGTGTAAAATCTTCAACATGTTTGTTGCTCCCAACAACACCAAGTCCAACTACTTTGGATTTTCAGCATCCTTTATCTGAAGAACTTATTGAGCAGCCCTTCCTTACATTAAATGGAGATAATATTGATATTGCTGACACCCAAAATGATGCTACAGCAGGTGATTCAAGTTCTTCGGCAGATTCTAACTCAGTTGTCCAGGATCCAGTTAGTGCACAATTGATAAACAATATTAGTATGTTAGATTATCAAACACTGAGTAAGAATGGGGATCTTATTATTGGACAGGCAGAAGATGTTAAACTGAATGGGAATTTGAAAATTAATAATAGGAAACTGCCAAGCTTTGTGAACTGGAACTGGGTTGTTATACGAAAGATTTTACTGTGGGTAGTACTGTCTGGGCTCATTGCTTGTCTTGCAGCTATCATTGGTATGATAATAACTATTCCTAAAGAATGTAATATTGACCTACCATGGTATCAAGGAAAGGTTTTTTACGAAGTATTTCCTGCCAGTTTTAAAGATTCCAATAATGATGGCACAGGTGATTTTAAAGGCCTGATAACTAAACTTGATTATATACAAAATTTAGGTGTTGCTGCTATTCGACTTAATTATATATTTCAAGCTGATCATTATCCTGAAGATTACAATAATGTAACTTCTATGTTGGACATAGACAGGAGCTTAGGCGTACTTAAAGACTTACAGGATCTAATATTTAATATACATAAAAGGAACATGTCAGTCATACTTGATTTACCAGTAATTTCAATGGCTCTTCCAAGTCTCATTTTTAACTCCACTTCATCTATTATCATAGCAAATAATACTGAAGTTAACTATTTGGATCCAACAACTGTTGCTATAATTCATTGGTCTCAAGCACAAGGAGTTGATGGATTTTATTTAAAAAAATTGGAAAATTTTGTGAACGAACCATATTTTGGTAGAACTCTTCAGATTTGGAAACAAATTATAGGATTCAATAAAATTTTGATTGCGAGCGAATTGGCTTATAGTAAAGCTACAGGTGATGCTTTGAATATTCTTTTGTCAAGAATTGATCTCATAGATGTTCACATAGATTTAGAAGAAGAAACTGCTGGTATTTGTGATAAAATCAAGAAGGTTGTGGATGGACAATTATGGTCTAAGCCTCATTATCCTTGGGTACATTGGAATATTGGTAATGTGAACAGTGAAAGAATAGCGACAAAACATATTAATAATACTTTAGCTTTGACAGTTTTAGAATTCGTTCTTCCAGGAACTGTAAGTATATTCTACGGAGATGAGATAGGATTGGGCGGTATACCAGATGATGTTGAAGAAGATTTCCATGAACACAAAGATATTCATAACTTAGTCCAAATGGCTTTTGCTGATAGTGAGGTGACCCCTACTAATGTTGAAATTCTACCATGGAACCTAAAGTCTGTACTAAAGCCAAATTATGATTTTCTTAATGTCATTAAAAACTTGATTAAGGTAAGAGTGACTACTCCTACAATTTACTTAAGAGCAATTTTCAAGGAGGGCGTTATGCAACCTAATGTAGAAATACGTCAAACGGAAGAAGATTTGATTGTGATTGAGCGTTGGTACCCTCGTAGAAATGCCTGCGTTTTTGTCGGTAATCTAGGCAGTAAGACTATAACAGCCGATTTATCTACAATGTTCTATGGTGGGAAAGTTATTGCTGGCACTAATTCTTCATTAATTGGTCAAGTATTGTATTTTGAGAAAGTGACTTTCTCGCCCAATTCAGCAATAATAGTTAAAATGGAGAAATAA
Protein
MDSLPENSTNGGSQLETSDYLHGVKSSTCLLLPTTPSPTTLDFQHPLSEELIEQPFLTLNGDNIDIADTQNDATAGDSSSSADSNSVVQDPVSAQLINNISMLDYQTLSKNGDLIIGQAEDVKLNGNLKINNRKLPSFVNWNWVVIRKILLWVVLSGLIACLAAIIGMIITIPKECNIDLPWYQGKVFYEVFPASFKDSNNDGTGDFKGLITKLDYIQNLGVAAIRLNYIFQADHYPEDYNNVTSMLDIDRSLGVLKDLQDLIFNIHKRNMSVILDLPVISMALPSLIFNSTSSIIIANNTEVNYLDPTTVAIIHWSQAQGVDGFYLKKLENFVNEPYFGRTLQIWKQIIGFNKILIASELAYSKATGDALNILLSRIDLIDVHIDLEEETAGICDKIKKVVDGQLWSKPHYPWVHWNIGNVNSERIATKHINNTLALTVLEFVLPGTVSIFYGDEIGLGGIPDDVEEDFHEHKDIHNLVQMAFADSEVTPTNVEILPWNLKSVLKPNYDFLNVIKNLIKVRVTTPTIYLRAIFKEGVMQPNVEIRQTEEDLIVIERWYPRRNACVFVGNLGSKTITADLSTMFYGGKVIAGTNSSLIGQVLYFEKVTFSPNSAIIVKMEK
Summary
Uniprot
A0A2H1V3W4
A0A2A4JZY3
A0A0L7LA37
A0A212FLL9
A0A194R124
A0A194PHF4
+ More
A0A1S4FJN7 Q16YM2 Q5TND7 A0A182KUL1 A0A182WYQ4 A0A023EVE8 A0A182I519 A0A182VIA3 A0A182SHS9 A0A182R9L7 A0A182QGC4 A0A084WBX7 U5EPQ3 A0A182K387 A0A182UC15 A0A182YFC1 A0A182NEZ8 A0A1B0CUJ4 A0A026VUK7 A0A0J7KHP8 A0A1I8M964 A0A182F5E2 T1PA19 W5JLN1 A0A182ITW5 A0A182WH20 B0W1I2 A0A195CMH7 A0A1Y1NH85 A0A158N9F8 A0A182MWR0 A0A151HZR7 A0A151JV08 A0A0L7RFU7 A0A088AAX5 A0A034WD52 A0A0K8VPR1 E2BFJ4 A0A0L0CBK5 A0A151WGR5 A0A0K8UEC7 A0A195E1E7 A0A0A1X050 A0A182NZV7 D6X352 A0A2A3EE62 A0A232F8F5 K7IWH6 A0A1Q3FM05 E2AMD2 A0A1I8PT10 E9IW30 A0A1I8PSW7 W8AVH0 A0A1A9W5C9 A0A0M9A678 A0A1W4XB62 A0A1B0FG52 A0A1B0A4R5 A0A1A9V2A0 A0A1B6H479 A0A0K8U294 E0VNW9 A0A067R5T3 A0A1B0ANZ1 A0A1A9XVU9 A0A2J7PKA0 A0A2J7PK94 A0A2J7PK91 A0A069DZF0 A0A1B6CHZ3 A0A023F646 A0A0V0G6M3 A0A0P4W495 T1HSS6 A0A154PE16 A0A0C9Q0L8 A0A0C9RLQ3 A0A0C9RGD8 A0A336M3K7 A0A1B0D366 A0A1W7R8J0 A0A2R7W383 A0A336KU16 A0A1B6MBZ4 A0A3Q0IYL9 A0A0Q9WHK9 A0A0Q9XJJ1 A0A0M3QTN2 A0A0P5X0U7 A0A0P5YPG8
A0A1S4FJN7 Q16YM2 Q5TND7 A0A182KUL1 A0A182WYQ4 A0A023EVE8 A0A182I519 A0A182VIA3 A0A182SHS9 A0A182R9L7 A0A182QGC4 A0A084WBX7 U5EPQ3 A0A182K387 A0A182UC15 A0A182YFC1 A0A182NEZ8 A0A1B0CUJ4 A0A026VUK7 A0A0J7KHP8 A0A1I8M964 A0A182F5E2 T1PA19 W5JLN1 A0A182ITW5 A0A182WH20 B0W1I2 A0A195CMH7 A0A1Y1NH85 A0A158N9F8 A0A182MWR0 A0A151HZR7 A0A151JV08 A0A0L7RFU7 A0A088AAX5 A0A034WD52 A0A0K8VPR1 E2BFJ4 A0A0L0CBK5 A0A151WGR5 A0A0K8UEC7 A0A195E1E7 A0A0A1X050 A0A182NZV7 D6X352 A0A2A3EE62 A0A232F8F5 K7IWH6 A0A1Q3FM05 E2AMD2 A0A1I8PT10 E9IW30 A0A1I8PSW7 W8AVH0 A0A1A9W5C9 A0A0M9A678 A0A1W4XB62 A0A1B0FG52 A0A1B0A4R5 A0A1A9V2A0 A0A1B6H479 A0A0K8U294 E0VNW9 A0A067R5T3 A0A1B0ANZ1 A0A1A9XVU9 A0A2J7PKA0 A0A2J7PK94 A0A2J7PK91 A0A069DZF0 A0A1B6CHZ3 A0A023F646 A0A0V0G6M3 A0A0P4W495 T1HSS6 A0A154PE16 A0A0C9Q0L8 A0A0C9RLQ3 A0A0C9RGD8 A0A336M3K7 A0A1B0D366 A0A1W7R8J0 A0A2R7W383 A0A336KU16 A0A1B6MBZ4 A0A3Q0IYL9 A0A0Q9WHK9 A0A0Q9XJJ1 A0A0M3QTN2 A0A0P5X0U7 A0A0P5YPG8
Pubmed
26227816
22118469
26354079
17510324
12364791
14747013
+ More
17210077 20966253 24945155 24438588 25244985 24508170 30249741 25315136 20920257 23761445 28004739 21347285 25348373 20798317 26108605 25830018 18362917 19820115 28648823 20075255 21282665 24495485 20566863 24845553 26334808 25474469 27129103 17994087 18057021
17210077 20966253 24945155 24438588 25244985 24508170 30249741 25315136 20920257 23761445 28004739 21347285 25348373 20798317 26108605 25830018 18362917 19820115 28648823 20075255 21282665 24495485 20566863 24845553 26334808 25474469 27129103 17994087 18057021
EMBL
ODYU01000563
SOQ35535.1
NWSH01000345
PCG77238.1
JTDY01002044
KOB72250.1
+ More
AGBW02007731 OWR54648.1 KQ460883 KPJ11214.1 KQ459603 KPI92742.1 CH477514 EAT39713.1 AAAB01008984 EAL39050.2 GAPW01000490 JAC13108.1 APCN01003748 AXCN02000838 ATLV01022515 KE525333 KFB47721.1 GANO01003626 JAB56245.1 AJWK01029249 KK107928 QOIP01000001 EZA47171.1 RLU26514.1 LBMM01007389 KMQ89777.1 KA645582 AFP60211.1 ADMH02000797 ETN65031.1 DS231822 EDS25780.1 KQ977565 KYN01923.1 GEZM01005656 JAV96005.1 ADTU01009632 AXCM01001497 KQ976712 KYM76973.1 KQ981713 KYN37379.1 KQ414601 KOC69720.1 GAKP01006716 GAKP01006715 JAC52237.1 GDHF01011754 JAI40560.1 GL448037 EFN85516.1 JRES01000648 KNC29610.1 KQ983152 KYQ47021.1 GDHF01027287 JAI25027.1 KQ979814 KYN18975.1 GBXI01010264 JAD04028.1 KQ971372 EFA09799.1 KZ288267 PBC30073.1 NNAY01000694 OXU26945.1 GFDL01006483 JAV28562.1 GL440811 EFN65423.1 GL766449 EFZ15201.1 GAMC01016273 GAMC01016271 JAB90282.1 KQ435742 KOX76753.1 CCAG010013157 GECZ01000278 JAS69491.1 GDHF01031450 JAI20864.1 DS235354 EEB15075.1 KK852723 KDR17684.1 JXJN01001085 NEVH01024940 PNF16755.1 PNF16754.1 PNF16753.1 GBGD01000755 JAC88134.1 GEDC01024305 GEDC01013378 JAS12993.1 JAS23920.1 GBBI01002030 JAC16682.1 GECL01003042 JAP03082.1 GDKW01000436 JAI56159.1 ACPB03004812 KQ434886 KZC10081.1 GBYB01007418 JAG77185.1 GBYB01009205 JAG78972.1 GBYB01007415 JAG77182.1 UFQS01000502 UFQT01000502 SSX04461.1 SSX24825.1 AJVK01023317 AJVK01023318 GEHC01000202 JAV47443.1 KK854262 PTY13888.1 SSX04460.1 SSX24824.1 GEBQ01006524 JAT33453.1 CH940649 KRF81687.1 KRF81688.1 CH933807 KRG03827.1 CP012523 ALC39179.1 GDIP01086498 GDIP01079243 JAM24472.1 GDIP01056293 JAM47422.1
AGBW02007731 OWR54648.1 KQ460883 KPJ11214.1 KQ459603 KPI92742.1 CH477514 EAT39713.1 AAAB01008984 EAL39050.2 GAPW01000490 JAC13108.1 APCN01003748 AXCN02000838 ATLV01022515 KE525333 KFB47721.1 GANO01003626 JAB56245.1 AJWK01029249 KK107928 QOIP01000001 EZA47171.1 RLU26514.1 LBMM01007389 KMQ89777.1 KA645582 AFP60211.1 ADMH02000797 ETN65031.1 DS231822 EDS25780.1 KQ977565 KYN01923.1 GEZM01005656 JAV96005.1 ADTU01009632 AXCM01001497 KQ976712 KYM76973.1 KQ981713 KYN37379.1 KQ414601 KOC69720.1 GAKP01006716 GAKP01006715 JAC52237.1 GDHF01011754 JAI40560.1 GL448037 EFN85516.1 JRES01000648 KNC29610.1 KQ983152 KYQ47021.1 GDHF01027287 JAI25027.1 KQ979814 KYN18975.1 GBXI01010264 JAD04028.1 KQ971372 EFA09799.1 KZ288267 PBC30073.1 NNAY01000694 OXU26945.1 GFDL01006483 JAV28562.1 GL440811 EFN65423.1 GL766449 EFZ15201.1 GAMC01016273 GAMC01016271 JAB90282.1 KQ435742 KOX76753.1 CCAG010013157 GECZ01000278 JAS69491.1 GDHF01031450 JAI20864.1 DS235354 EEB15075.1 KK852723 KDR17684.1 JXJN01001085 NEVH01024940 PNF16755.1 PNF16754.1 PNF16753.1 GBGD01000755 JAC88134.1 GEDC01024305 GEDC01013378 JAS12993.1 JAS23920.1 GBBI01002030 JAC16682.1 GECL01003042 JAP03082.1 GDKW01000436 JAI56159.1 ACPB03004812 KQ434886 KZC10081.1 GBYB01007418 JAG77185.1 GBYB01009205 JAG78972.1 GBYB01007415 JAG77182.1 UFQS01000502 UFQT01000502 SSX04461.1 SSX24825.1 AJVK01023317 AJVK01023318 GEHC01000202 JAV47443.1 KK854262 PTY13888.1 SSX04460.1 SSX24824.1 GEBQ01006524 JAT33453.1 CH940649 KRF81687.1 KRF81688.1 CH933807 KRG03827.1 CP012523 ALC39179.1 GDIP01086498 GDIP01079243 JAM24472.1 GDIP01056293 JAM47422.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
UP000008820
+ More
UP000007062 UP000075882 UP000076407 UP000075840 UP000075903 UP000075901 UP000075900 UP000075886 UP000030765 UP000075881 UP000075902 UP000076408 UP000075884 UP000092461 UP000053097 UP000279307 UP000036403 UP000095301 UP000069272 UP000000673 UP000075880 UP000075920 UP000002320 UP000078542 UP000005205 UP000075883 UP000078540 UP000078541 UP000053825 UP000005203 UP000008237 UP000037069 UP000075809 UP000078492 UP000075885 UP000007266 UP000242457 UP000215335 UP000002358 UP000000311 UP000095300 UP000091820 UP000053105 UP000192223 UP000092444 UP000092445 UP000078200 UP000009046 UP000027135 UP000092460 UP000092443 UP000235965 UP000015103 UP000076502 UP000092462 UP000079169 UP000008792 UP000009192 UP000092553
UP000007062 UP000075882 UP000076407 UP000075840 UP000075903 UP000075901 UP000075900 UP000075886 UP000030765 UP000075881 UP000075902 UP000076408 UP000075884 UP000092461 UP000053097 UP000279307 UP000036403 UP000095301 UP000069272 UP000000673 UP000075880 UP000075920 UP000002320 UP000078542 UP000005205 UP000075883 UP000078540 UP000078541 UP000053825 UP000005203 UP000008237 UP000037069 UP000075809 UP000078492 UP000075885 UP000007266 UP000242457 UP000215335 UP000002358 UP000000311 UP000095300 UP000091820 UP000053105 UP000192223 UP000092444 UP000092445 UP000078200 UP000009046 UP000027135 UP000092460 UP000092443 UP000235965 UP000015103 UP000076502 UP000092462 UP000079169 UP000008792 UP000009192 UP000092553
Pfam
PF00128 Alpha-amylase
SUPFAM
SSF51445
SSF51445
ProteinModelPortal
A0A2H1V3W4
A0A2A4JZY3
A0A0L7LA37
A0A212FLL9
A0A194R124
A0A194PHF4
+ More
A0A1S4FJN7 Q16YM2 Q5TND7 A0A182KUL1 A0A182WYQ4 A0A023EVE8 A0A182I519 A0A182VIA3 A0A182SHS9 A0A182R9L7 A0A182QGC4 A0A084WBX7 U5EPQ3 A0A182K387 A0A182UC15 A0A182YFC1 A0A182NEZ8 A0A1B0CUJ4 A0A026VUK7 A0A0J7KHP8 A0A1I8M964 A0A182F5E2 T1PA19 W5JLN1 A0A182ITW5 A0A182WH20 B0W1I2 A0A195CMH7 A0A1Y1NH85 A0A158N9F8 A0A182MWR0 A0A151HZR7 A0A151JV08 A0A0L7RFU7 A0A088AAX5 A0A034WD52 A0A0K8VPR1 E2BFJ4 A0A0L0CBK5 A0A151WGR5 A0A0K8UEC7 A0A195E1E7 A0A0A1X050 A0A182NZV7 D6X352 A0A2A3EE62 A0A232F8F5 K7IWH6 A0A1Q3FM05 E2AMD2 A0A1I8PT10 E9IW30 A0A1I8PSW7 W8AVH0 A0A1A9W5C9 A0A0M9A678 A0A1W4XB62 A0A1B0FG52 A0A1B0A4R5 A0A1A9V2A0 A0A1B6H479 A0A0K8U294 E0VNW9 A0A067R5T3 A0A1B0ANZ1 A0A1A9XVU9 A0A2J7PKA0 A0A2J7PK94 A0A2J7PK91 A0A069DZF0 A0A1B6CHZ3 A0A023F646 A0A0V0G6M3 A0A0P4W495 T1HSS6 A0A154PE16 A0A0C9Q0L8 A0A0C9RLQ3 A0A0C9RGD8 A0A336M3K7 A0A1B0D366 A0A1W7R8J0 A0A2R7W383 A0A336KU16 A0A1B6MBZ4 A0A3Q0IYL9 A0A0Q9WHK9 A0A0Q9XJJ1 A0A0M3QTN2 A0A0P5X0U7 A0A0P5YPG8
A0A1S4FJN7 Q16YM2 Q5TND7 A0A182KUL1 A0A182WYQ4 A0A023EVE8 A0A182I519 A0A182VIA3 A0A182SHS9 A0A182R9L7 A0A182QGC4 A0A084WBX7 U5EPQ3 A0A182K387 A0A182UC15 A0A182YFC1 A0A182NEZ8 A0A1B0CUJ4 A0A026VUK7 A0A0J7KHP8 A0A1I8M964 A0A182F5E2 T1PA19 W5JLN1 A0A182ITW5 A0A182WH20 B0W1I2 A0A195CMH7 A0A1Y1NH85 A0A158N9F8 A0A182MWR0 A0A151HZR7 A0A151JV08 A0A0L7RFU7 A0A088AAX5 A0A034WD52 A0A0K8VPR1 E2BFJ4 A0A0L0CBK5 A0A151WGR5 A0A0K8UEC7 A0A195E1E7 A0A0A1X050 A0A182NZV7 D6X352 A0A2A3EE62 A0A232F8F5 K7IWH6 A0A1Q3FM05 E2AMD2 A0A1I8PT10 E9IW30 A0A1I8PSW7 W8AVH0 A0A1A9W5C9 A0A0M9A678 A0A1W4XB62 A0A1B0FG52 A0A1B0A4R5 A0A1A9V2A0 A0A1B6H479 A0A0K8U294 E0VNW9 A0A067R5T3 A0A1B0ANZ1 A0A1A9XVU9 A0A2J7PKA0 A0A2J7PK94 A0A2J7PK91 A0A069DZF0 A0A1B6CHZ3 A0A023F646 A0A0V0G6M3 A0A0P4W495 T1HSS6 A0A154PE16 A0A0C9Q0L8 A0A0C9RLQ3 A0A0C9RGD8 A0A336M3K7 A0A1B0D366 A0A1W7R8J0 A0A2R7W383 A0A336KU16 A0A1B6MBZ4 A0A3Q0IYL9 A0A0Q9WHK9 A0A0Q9XJJ1 A0A0M3QTN2 A0A0P5X0U7 A0A0P5YPG8
PDB
4GIN
E-value=3.74491e-16,
Score=209
Ontologies
GO
Topology
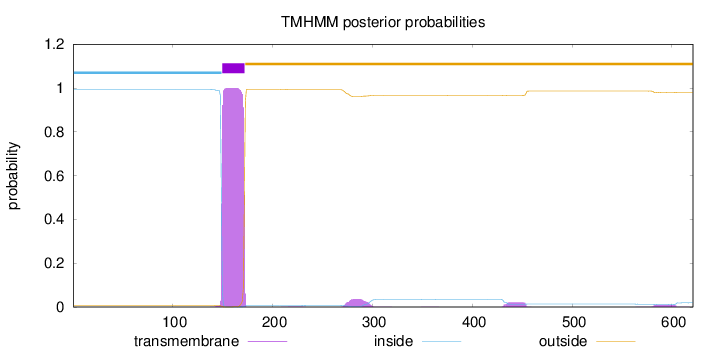
Length:
621
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.33504
Exp number, first 60 AAs:
0.00087
Total prob of N-in:
0.99326
inside
1 - 149
TMhelix
150 - 172
outside
173 - 621
Population Genetic Test Statistics
Pi
246.556718
Theta
225.894184
Tajima's D
0.621969
CLR
0
CSRT
0.550972451377431
Interpretation
Uncertain
