Gene
KWMTBOMO07167
Pre Gene Modal
BGIBMGA010557
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.786
Sequence
CDS
ATGAACATCGAGATGGGAAGTGCGTCCCAGGAATGCGGACGAGTAAAGGCTATTACTTTTTTGGATTTGGGATACAAAACCACAAATGCGCAAGTTTCAGGATTTCCGCTATGTTTCAATCGTAAATCCAGTACAACCTTAACTATTATTAATGGAGCCTGTGGAGTGATAAGGCCAGGACGCCTAACCTTCATTCTTGGACCCTCCGGTGCTGGAAAAACCACTCTTCTCAAAATAATAGCCAAACGAAAAAGATCCGGAGTAACGGGTTCCGTTTTTGGTGCCAACCGTAACGTTGTTTTTGTAGATCAACACACCACCCTAATAGAAACACTTACCGCAAGAGAAACTATAAAGTTCGCCGCGAGCCTAAAACTGGCGAAAATGAACTACCGAGAACGTATACAAATGGTGGAATCAGTAACAAAACAGTTGGGCATTTACGACATTCTTAACACAAAGTGCAGTCGTCTTTCGGGTGGAGAAAGGAAAAGACTTACAATCGCCTGTGAATTATTAACAGACCCTCCGATTATGCTTCTTGATGAACCGACGAGCGGCCTTGATACAGTGTCGTCATTGTCTGTGGTTCGAGCGCTACAGACAGTGGCACGCACTGGACGGATCGTGGCCTGCGTGGTACACCAACCTTCCTCACAACTATACGCAACAGCAGACGATGTCCTATTATTGGCCAACGGAAAAACCCTTTATGCTGGGCCACTGAGAGATATTCCTGGGACACTATTAAAAGCTGGCTTATCATGTCCACAATATTATAACATGGCTGATTATTTATTAGAGGTCGCAAGTTTAAATCATCCATCCCATCAAATTCTTGAAAGTGAAGCACGGAGCTATGCTATTGAATTGAGAAAAAATACACAAATCGACATTTCAATAAAAAATGGAGAAGAATCCTCACCAGAATCTGAAGCCTTACTAAATCCGTATCCTGTTAAGGATTCGCATACGTACACCGCAAATTCATTACAACAATGTAAAGCGCTACTGTGGAGATGCTTCATTGGAGCCGTCAGAGACGTACACATAACTCAGATTAGACTGGCCACCCATCTCGTAGTGGCGCTATTGTTAGGTGCACTATACAACAAAGCTGGCCTGGAAGCACACAGAATTATATCTAATACTGGATGTTTGTTTTTCTTTCTACTATTCTTATTCTTCTCTAATGCTATGCCCACTATTCATACGTTTCCATCAGAATCAACAGTCGTTCTTCAGCAGCATATGAACAGATGGTACTCCTTAGTTCTTTATTGTGGTTCGAAGATTATAGTAGATCTACCCATACAACTCCTTTGTTCCACTGTGTTCCTAATTCCAGCATGGTACTTAACATCGCAACCTACAGACTTATTTAGAGTAGGAATGGCGTGGACAATATGCGTACTCATGACTATTCTGGCCCAAACATTTGGTTTGGTAGTCGGTGCTGCTTATGGAATGAAGCTCGGACTGTTCATCATTCCCGCTGCAAACATTCCGATGCTGATGTTCTCAGAGTTTTTCATTCCGTACAAGGAAATACCGTCGTACCTTCAACCGCTCTGCGTGATCTCTTACTTTCGTTACGGTTTCAATGCCTTCCTTAAAATAGTATACGGTTATGGAAGAGAAAAACTACCCTGTCATGTAGTATTTTGCATGTTTAAGAATCCAGCCAAATATCTAGAATATTTGGGGATCTCTGATAATGTGACTTATGATTTTCTTGCTTTAGTAATCTGGATTATTGTTTTACAGATTACCTTAGTTTTTGTTCTTATGTTTAAGGCATATAATGCGTGCAGATGA
Protein
MNIEMGSASQECGRVKAITFLDLGYKTTNAQVSGFPLCFNRKSSTTLTIINGACGVIRPGRLTFILGPSGAGKTTLLKIIAKRKRSGVTGSVFGANRNVVFVDQHTTLIETLTARETIKFAASLKLAKMNYRERIQMVESVTKQLGIYDILNTKCSRLSGGERKRLTIACELLTDPPIMLLDEPTSGLDTVSSLSVVRALQTVARTGRIVACVVHQPSSQLYATADDVLLLANGKTLYAGPLRDIPGTLLKAGLSCPQYYNMADYLLEVASLNHPSHQILESEARSYAIELRKNTQIDISIKNGEESSPESEALLNPYPVKDSHTYTANSLQQCKALLWRCFIGAVRDVHITQIRLATHLVVALLLGALYNKAGLEAHRIISNTGCLFFFLLFLFFSNAMPTIHTFPSESTVVLQQHMNRWYSLVLYCGSKIIVDLPIQLLCSTVFLIPAWYLTSQPTDLFRVGMAWTICVLMTILAQTFGLVVGAAYGMKLGLFIIPAANIPMLMFSEFFIPYKEIPSYLQPLCVISYFRYGFNAFLKIVYGYGREKLPCHVVFCMFKNPAKYLEYLGISDNVTYDFLALVIWIIVLQITLVFVLMFKAYNACR
Summary
Uniprot
A0A212EZA6
A0A2H1V3X0
A0A194PJ41
A0A194R0I9
A0A2J7PQV5
A0A1L8E487
+ More
A0A2J7RQG6 A0A0L7QXB8 A0A154P0I1 U5ERX5 A0A0L0CCK3 A0A1A9WZY6 B4MZA9 A0A1A9XZH7 A0A1B0B920 A0A0Q9X319 A0A0Q9WIH6 A0A1Q3FIE6 B4LSY3 U5EQY5 A0A1Q3FI78 Q16Y91 A0A1I8PCP7 A0A158P162 A0A154PC74 W5JX33 T1PC81 A0A1S4FK05 A0A151X0G7 A0A182GYW0 A0A1A9UMH5 A0A182QY45 E2A2I5 Q16Y89 A0A1B0FIL4 A0A088A0P2 A0A151K005 A0A182S5B1 A0A1S4FJZ4 A0A158P3Y5 A0A1Q3FP51 A0A182INJ2 A0A026WFK9 A0A232F0W1 B0X147 A0A0N0U4H6 K7ILU9 B3MNY4 A0A182VHN3 A0A0N8NZB1 A0A1Y9GLC8 A0A2M3Z4N9 E2A627 A0A182L5H9 A0A195FTJ6 A0A151J3S1 A0A1S4HFH3 A0A182HF72 A0A182S5B0 B4JCD0 A0A182U9V7 B4KEP7 A0A1Y9GLZ1 Q7PFN4 A0A182UXV9 A0A182UHZ9 A0A1S4H190 E2C0H1 A0A2M4CMQ3 A0A0L7RDS6 A0A1Y9GKW8 A0A0C9S320 V9ILI0 A0A2A3EEV9 A0A0A9W0J1 A0A0C9RP34 A0A2M4BIA8 A0A151WQM4 F4W5H8 A0A195BKY4 A0A158NYX8 A0A088AN61 W5JV37 A0A161AM50 A0A182FWY0 K7IQL0 A0A146LKB6 A0A1B6F7Y9 B4NXU2 A0A182JJT5 A0A146LYU6 A0A3L8DX23 A0A182MEC7 A0A0R1DQ54 A0A182F589 A0A182YMF2 A0A182K2C9 A0A195CNC3 A0A3B0K836 V5I802
A0A2J7RQG6 A0A0L7QXB8 A0A154P0I1 U5ERX5 A0A0L0CCK3 A0A1A9WZY6 B4MZA9 A0A1A9XZH7 A0A1B0B920 A0A0Q9X319 A0A0Q9WIH6 A0A1Q3FIE6 B4LSY3 U5EQY5 A0A1Q3FI78 Q16Y91 A0A1I8PCP7 A0A158P162 A0A154PC74 W5JX33 T1PC81 A0A1S4FK05 A0A151X0G7 A0A182GYW0 A0A1A9UMH5 A0A182QY45 E2A2I5 Q16Y89 A0A1B0FIL4 A0A088A0P2 A0A151K005 A0A182S5B1 A0A1S4FJZ4 A0A158P3Y5 A0A1Q3FP51 A0A182INJ2 A0A026WFK9 A0A232F0W1 B0X147 A0A0N0U4H6 K7ILU9 B3MNY4 A0A182VHN3 A0A0N8NZB1 A0A1Y9GLC8 A0A2M3Z4N9 E2A627 A0A182L5H9 A0A195FTJ6 A0A151J3S1 A0A1S4HFH3 A0A182HF72 A0A182S5B0 B4JCD0 A0A182U9V7 B4KEP7 A0A1Y9GLZ1 Q7PFN4 A0A182UXV9 A0A182UHZ9 A0A1S4H190 E2C0H1 A0A2M4CMQ3 A0A0L7RDS6 A0A1Y9GKW8 A0A0C9S320 V9ILI0 A0A2A3EEV9 A0A0A9W0J1 A0A0C9RP34 A0A2M4BIA8 A0A151WQM4 F4W5H8 A0A195BKY4 A0A158NYX8 A0A088AN61 W5JV37 A0A161AM50 A0A182FWY0 K7IQL0 A0A146LKB6 A0A1B6F7Y9 B4NXU2 A0A182JJT5 A0A146LYU6 A0A3L8DX23 A0A182MEC7 A0A0R1DQ54 A0A182F589 A0A182YMF2 A0A182K2C9 A0A195CNC3 A0A3B0K836 V5I802
Pubmed
EMBL
AGBW02011356
OWR46791.1
ODYU01000563
SOQ35538.1
KQ459603
KPI92739.1
+ More
KQ460883 KPJ11217.1 NEVH01022635 PNF18686.1 GFDF01000537 JAV13547.1 NEVH01001337 PNF43079.1 KQ414705 KOC63206.1 KQ434783 KZC04864.1 GANO01002627 JAB57244.1 JRES01000682 KNC29189.1 CH963913 EDW77382.2 JXJN01010202 KRF98663.1 CH940649 KRF81927.1 GFDL01007717 JAV27328.1 EDW64894.1 GANO01003058 JAB56813.1 GFDL01007797 JAV27248.1 CH477522 EAT39577.1 ADTU01005820 ADTU01005821 ADTU01005822 KQ434864 KZC09024.1 ADMH02000042 ETN68005.1 KA646371 AFP61000.1 KQ982612 KYQ53870.1 JXUM01021316 JXUM01021317 JXUM01021318 JXUM01021319 JXUM01021320 JXUM01021321 JXUM01021322 JXUM01021323 AXCN02000853 GL436038 EFN72388.1 EAT39579.1 CCAG010016358 KQ981414 KYN41880.1 ADTU01008610 ADTU01008611 GFDL01005680 JAV29365.1 KK107235 QOIP01000009 EZA54900.1 RLU19114.1 NNAY01001302 OXU24426.1 DS232252 EDS38496.1 KQ435824 KOX72124.1 CH902620 EDV32171.1 KPU73827.1 KPU73828.1 APCN01004583 GGFM01002714 MBW23465.1 GL437108 EFN71086.1 KQ981276 KYN43612.1 KQ980249 KYN17092.1 AAAB01008839 JXUM01036537 JXUM01036538 JXUM01036539 JXUM01036540 KQ561097 KXJ79717.1 CH916368 EDW04163.1 CH933807 EDW12947.1 KRG03515.1 KRG03516.1 EAA45245.4 GL451783 EFN78564.1 GGFL01002428 MBW66606.1 KQ414613 KOC69008.1 GBYB01015296 JAG85063.1 JR053625 AEY61978.1 KZ288269 PBC30014.1 GBHO01041657 JAG01947.1 GBYB01015295 GBYB01015297 JAG85062.1 JAG85064.1 GGFJ01003407 MBW52548.1 KQ982821 KYQ50166.1 GL887645 EGI70628.1 KQ976453 KYM85342.1 ADTU01004385 ETN68006.1 KF828802 AIN44119.1 GDHC01011333 JAQ07296.1 GECZ01023427 JAS46342.1 CM000157 EDW87513.1 KRJ97185.1 KRJ97186.1 GDHC01005868 JAQ12761.1 QOIP01000003 RLU24926.1 AXCM01001117 KRJ97184.1 KQ977513 KYN02165.1 OUUW01000006 SPP81796.1 GALX01005540 JAB62926.1
KQ460883 KPJ11217.1 NEVH01022635 PNF18686.1 GFDF01000537 JAV13547.1 NEVH01001337 PNF43079.1 KQ414705 KOC63206.1 KQ434783 KZC04864.1 GANO01002627 JAB57244.1 JRES01000682 KNC29189.1 CH963913 EDW77382.2 JXJN01010202 KRF98663.1 CH940649 KRF81927.1 GFDL01007717 JAV27328.1 EDW64894.1 GANO01003058 JAB56813.1 GFDL01007797 JAV27248.1 CH477522 EAT39577.1 ADTU01005820 ADTU01005821 ADTU01005822 KQ434864 KZC09024.1 ADMH02000042 ETN68005.1 KA646371 AFP61000.1 KQ982612 KYQ53870.1 JXUM01021316 JXUM01021317 JXUM01021318 JXUM01021319 JXUM01021320 JXUM01021321 JXUM01021322 JXUM01021323 AXCN02000853 GL436038 EFN72388.1 EAT39579.1 CCAG010016358 KQ981414 KYN41880.1 ADTU01008610 ADTU01008611 GFDL01005680 JAV29365.1 KK107235 QOIP01000009 EZA54900.1 RLU19114.1 NNAY01001302 OXU24426.1 DS232252 EDS38496.1 KQ435824 KOX72124.1 CH902620 EDV32171.1 KPU73827.1 KPU73828.1 APCN01004583 GGFM01002714 MBW23465.1 GL437108 EFN71086.1 KQ981276 KYN43612.1 KQ980249 KYN17092.1 AAAB01008839 JXUM01036537 JXUM01036538 JXUM01036539 JXUM01036540 KQ561097 KXJ79717.1 CH916368 EDW04163.1 CH933807 EDW12947.1 KRG03515.1 KRG03516.1 EAA45245.4 GL451783 EFN78564.1 GGFL01002428 MBW66606.1 KQ414613 KOC69008.1 GBYB01015296 JAG85063.1 JR053625 AEY61978.1 KZ288269 PBC30014.1 GBHO01041657 JAG01947.1 GBYB01015295 GBYB01015297 JAG85062.1 JAG85064.1 GGFJ01003407 MBW52548.1 KQ982821 KYQ50166.1 GL887645 EGI70628.1 KQ976453 KYM85342.1 ADTU01004385 ETN68006.1 KF828802 AIN44119.1 GDHC01011333 JAQ07296.1 GECZ01023427 JAS46342.1 CM000157 EDW87513.1 KRJ97185.1 KRJ97186.1 GDHC01005868 JAQ12761.1 QOIP01000003 RLU24926.1 AXCM01001117 KRJ97184.1 KQ977513 KYN02165.1 OUUW01000006 SPP81796.1 GALX01005540 JAB62926.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000235965
UP000053825
UP000076502
+ More
UP000037069 UP000091820 UP000007798 UP000092443 UP000092460 UP000008792 UP000008820 UP000095300 UP000005205 UP000000673 UP000095301 UP000075809 UP000069940 UP000078200 UP000075886 UP000000311 UP000092444 UP000005203 UP000078541 UP000075900 UP000075880 UP000053097 UP000279307 UP000215335 UP000002320 UP000053105 UP000002358 UP000007801 UP000075903 UP000075840 UP000075882 UP000078492 UP000249989 UP000001070 UP000075902 UP000009192 UP000007062 UP000008237 UP000242457 UP000007755 UP000078540 UP000069272 UP000002282 UP000075883 UP000076408 UP000075881 UP000078542 UP000268350
UP000037069 UP000091820 UP000007798 UP000092443 UP000092460 UP000008792 UP000008820 UP000095300 UP000005205 UP000000673 UP000095301 UP000075809 UP000069940 UP000078200 UP000075886 UP000000311 UP000092444 UP000005203 UP000078541 UP000075900 UP000075880 UP000053097 UP000279307 UP000215335 UP000002320 UP000053105 UP000002358 UP000007801 UP000075903 UP000075840 UP000075882 UP000078492 UP000249989 UP000001070 UP000075902 UP000009192 UP000007062 UP000008237 UP000242457 UP000007755 UP000078540 UP000069272 UP000002282 UP000075883 UP000076408 UP000075881 UP000078542 UP000268350
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A212EZA6
A0A2H1V3X0
A0A194PJ41
A0A194R0I9
A0A2J7PQV5
A0A1L8E487
+ More
A0A2J7RQG6 A0A0L7QXB8 A0A154P0I1 U5ERX5 A0A0L0CCK3 A0A1A9WZY6 B4MZA9 A0A1A9XZH7 A0A1B0B920 A0A0Q9X319 A0A0Q9WIH6 A0A1Q3FIE6 B4LSY3 U5EQY5 A0A1Q3FI78 Q16Y91 A0A1I8PCP7 A0A158P162 A0A154PC74 W5JX33 T1PC81 A0A1S4FK05 A0A151X0G7 A0A182GYW0 A0A1A9UMH5 A0A182QY45 E2A2I5 Q16Y89 A0A1B0FIL4 A0A088A0P2 A0A151K005 A0A182S5B1 A0A1S4FJZ4 A0A158P3Y5 A0A1Q3FP51 A0A182INJ2 A0A026WFK9 A0A232F0W1 B0X147 A0A0N0U4H6 K7ILU9 B3MNY4 A0A182VHN3 A0A0N8NZB1 A0A1Y9GLC8 A0A2M3Z4N9 E2A627 A0A182L5H9 A0A195FTJ6 A0A151J3S1 A0A1S4HFH3 A0A182HF72 A0A182S5B0 B4JCD0 A0A182U9V7 B4KEP7 A0A1Y9GLZ1 Q7PFN4 A0A182UXV9 A0A182UHZ9 A0A1S4H190 E2C0H1 A0A2M4CMQ3 A0A0L7RDS6 A0A1Y9GKW8 A0A0C9S320 V9ILI0 A0A2A3EEV9 A0A0A9W0J1 A0A0C9RP34 A0A2M4BIA8 A0A151WQM4 F4W5H8 A0A195BKY4 A0A158NYX8 A0A088AN61 W5JV37 A0A161AM50 A0A182FWY0 K7IQL0 A0A146LKB6 A0A1B6F7Y9 B4NXU2 A0A182JJT5 A0A146LYU6 A0A3L8DX23 A0A182MEC7 A0A0R1DQ54 A0A182F589 A0A182YMF2 A0A182K2C9 A0A195CNC3 A0A3B0K836 V5I802
A0A2J7RQG6 A0A0L7QXB8 A0A154P0I1 U5ERX5 A0A0L0CCK3 A0A1A9WZY6 B4MZA9 A0A1A9XZH7 A0A1B0B920 A0A0Q9X319 A0A0Q9WIH6 A0A1Q3FIE6 B4LSY3 U5EQY5 A0A1Q3FI78 Q16Y91 A0A1I8PCP7 A0A158P162 A0A154PC74 W5JX33 T1PC81 A0A1S4FK05 A0A151X0G7 A0A182GYW0 A0A1A9UMH5 A0A182QY45 E2A2I5 Q16Y89 A0A1B0FIL4 A0A088A0P2 A0A151K005 A0A182S5B1 A0A1S4FJZ4 A0A158P3Y5 A0A1Q3FP51 A0A182INJ2 A0A026WFK9 A0A232F0W1 B0X147 A0A0N0U4H6 K7ILU9 B3MNY4 A0A182VHN3 A0A0N8NZB1 A0A1Y9GLC8 A0A2M3Z4N9 E2A627 A0A182L5H9 A0A195FTJ6 A0A151J3S1 A0A1S4HFH3 A0A182HF72 A0A182S5B0 B4JCD0 A0A182U9V7 B4KEP7 A0A1Y9GLZ1 Q7PFN4 A0A182UXV9 A0A182UHZ9 A0A1S4H190 E2C0H1 A0A2M4CMQ3 A0A0L7RDS6 A0A1Y9GKW8 A0A0C9S320 V9ILI0 A0A2A3EEV9 A0A0A9W0J1 A0A0C9RP34 A0A2M4BIA8 A0A151WQM4 F4W5H8 A0A195BKY4 A0A158NYX8 A0A088AN61 W5JV37 A0A161AM50 A0A182FWY0 K7IQL0 A0A146LKB6 A0A1B6F7Y9 B4NXU2 A0A182JJT5 A0A146LYU6 A0A3L8DX23 A0A182MEC7 A0A0R1DQ54 A0A182F589 A0A182YMF2 A0A182K2C9 A0A195CNC3 A0A3B0K836 V5I802
PDB
6FFC
E-value=6.55602e-46,
Score=466
Ontologies
GO
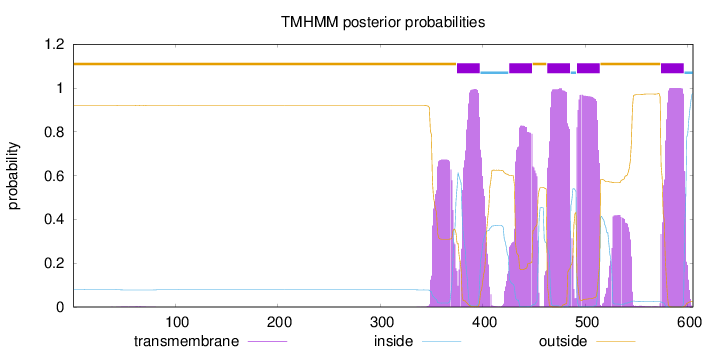
Topology
Length:
605
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
129.23265
Exp number, first 60 AAs:
0.032
Total prob of N-in:
0.07936
outside
1 - 374
TMhelix
375 - 397
inside
398 - 425
TMhelix
426 - 448
outside
449 - 462
TMhelix
463 - 485
inside
486 - 491
TMhelix
492 - 514
outside
515 - 573
TMhelix
574 - 596
inside
597 - 605
Population Genetic Test Statistics
Pi
164.566995
Theta
159.826986
Tajima's D
-0.817975
CLR
0.205709
CSRT
0.175291235438228
Interpretation
Uncertain
